MiFuP
MiFuP (Microbial Functional Potential) is a database of functional potentials deduced from microbial genomes.

Background
Microbial genome sequences data have been increased under next-generation sequencing technology in recent years.
Many useful functions and enzymes have been discovered from microbes, and industrially-utilized. For more effective industrial-utilization of microbial functions, a handy tool for searching enzymes and predicting microbial functions from genome sequence is necessary.
Summary of MiFuP
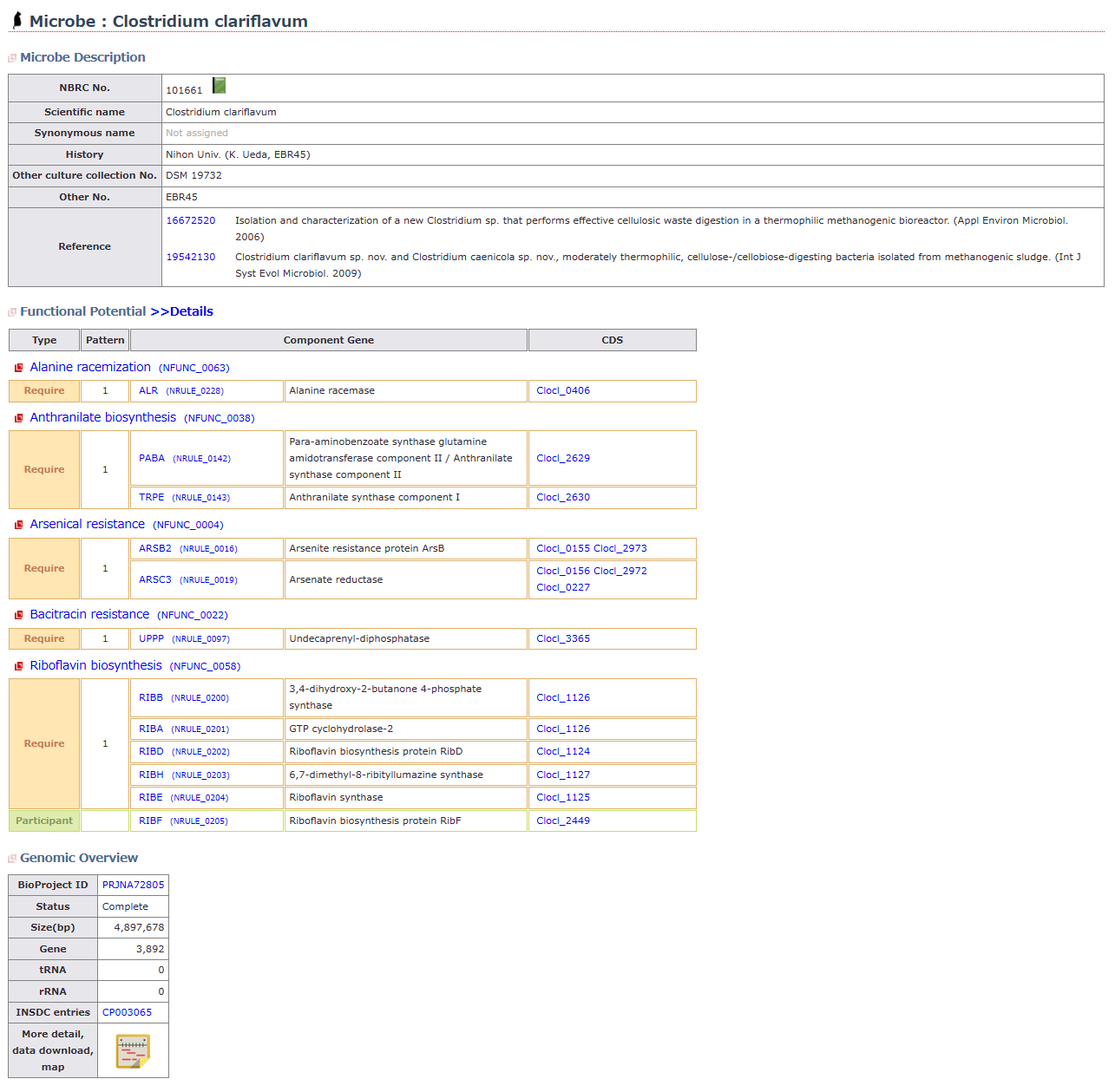
MiFuP is a database of functional potentials deduced from microbial genomes.
Each function is defined by annotators specialized in microbial field based on published reference. Key enzymes necessary for exertion of function, and threshold acquiring its orthologs are also defined.
- You can easily search for existing microbes with potential functions of your interest.
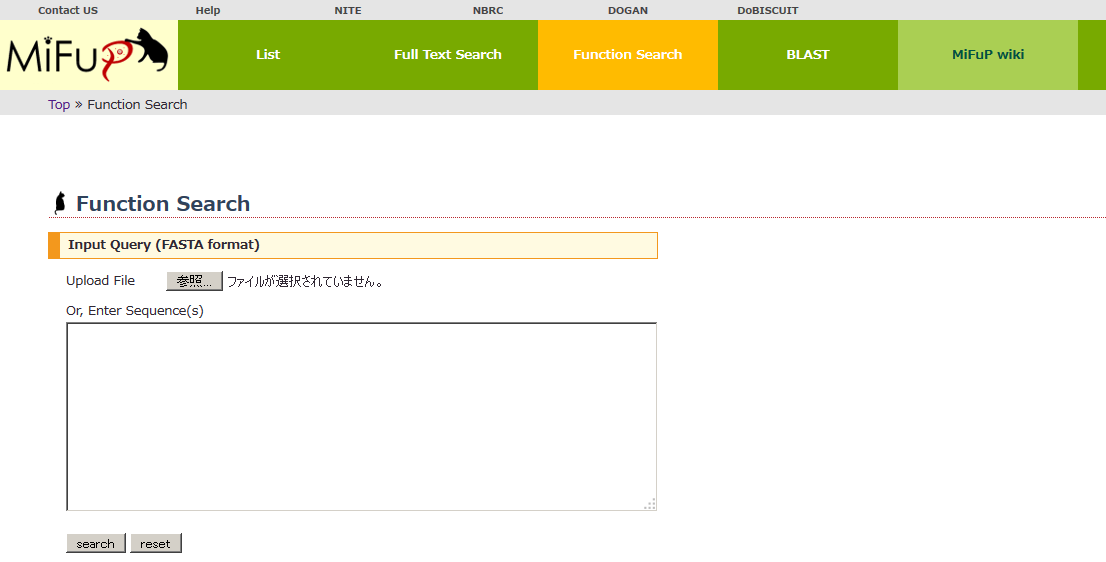
- You can search for functional potentials from your own microbial genome sequences or CDS sequences.
59 reference genomes were newly added (Dec. 15, 2016)
59 new reference genomes were added to the MiFuP database.
This database currently covers 66 bacterial and 9 archaeal classes and provides 355 strains (344 species) data.
http://www.bio.nite.go.jp/mifup/news
Contact us
- National Institute of Technology and Evaluation