Rifamycin : Cluster information
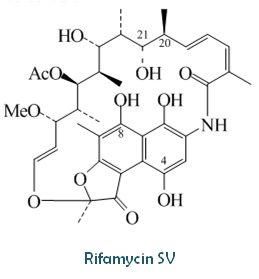
 Compound
Compound
| Entry name | Rifamycin |
|---|---|
| PKS Type | TypeI modular |
| Starter Unit | 3-amino-5-hydroxybenzoic acid (AHBA) |
| Chain Length | 11 |
| Sugar Unit | none |
| Classification | Ansamycin |
| Activity | Antibacterial |
| Composition | SV: C37H47NO12 |
 Original source
Original source
| Organism | Amycolatopsis mediterranei |
|---|---|
| Strain | S699 |
| Contig | AF040570 |
 PKS/NRPS Module
PKS/NRPS Module
| Rifam_00210 rifA | 0 |  | |
| 1 |  | methylmalonyl-CoA | |
| 2 |  | malonyl-CoA | |
| 3 |  | methylmalonyl-CoA | |
| Rifam_00220 rifB | 4 |  | methylmalonyl-CoA |
| 5 |  | methylmalonyl-CoA | |
| 6 |  | methylmalonyl-CoA | |
| Rifam_00230 rifC | 7 |  | methylmalonyl-CoA |
| Rifam_00240 rifD | 8 |  | methylmalonyl-CoA |
| Rifam_00250 rifE | 9 |  | malonyl-CoA |
| 10 |  | methylmalonyl-CoA |
 Reference
Reference
- 3-Amino-5-hydroxybenzoic acid synthase, the terminal enzyme in the formation of the precursor of mC7N units in rifamycin and related antibiotics.
- Kim CG, Yu TW, Fryhle CB, Handa S, Floss HG[PMID: 9497318]J Biol Chem. 273 (1998) 6030-40
- Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699.
- August PR, Tang L, Yoon YJ, Ning S, Muller R, Yu TW, Taylor M, Hoffmann D, Kim CG, Zhang X, Hutchinson CR, Floss HG[PMID: 9512878]Chem Biol. 5 (1998) 69-79
- Direct evidence that the rifamycin polyketide synthase assembles polyketide chains processively.
- Yu TW, Shen Y, Doi-Katayama Y, Tang L, Park C, Moore BS, Richard Hutchinson C, Floss HG[PMID: 10430893]Proc Natl Acad Sci U S A. 96 (1999) 9051-6
- Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase.
- Eads JC, Beeby M, Scapin G, Yu TW, Floss HG[PMID: 10433690]Biochemistry. 38 (1999) 9840-9
- Thioesterases and the premature termination of polyketide chain elongation in rifamycin B biosynthesis by Amycolatopsis mediterranei S699.
- Doi-Katayama Y, Yoon YJ, Choi CY, Yu TW, Floss HG, Hutchinson CR[PMID: 10908112]J Antibiot (Tokyo). 53 (2000) 484-95
- Mutational analysis and reconstituted expression of the biosynthetic genes involved in the formation of 3-amino-5-hydroxybenzoic acid, the starter unit of rifamycin biosynthesis in amycolatopsis Mediterranei S699.
- Yu TW, Muller R, Muller M, Zhang X, Draeger G, Kim CG, Leistner E, Floss HG[PMID: 11278540]J Biol Chem. 276 (2001) 12546-55
- The loading module of rifamycin synthetase is an adenylation-thiolation didomain with substrate tolerance for substituted benzoates.
- Admiraal SJ, Walsh CT, Khosla C[PMID: 11352749]Biochemistry. 40 (2001) 6116-23
- Biosynthesis of 1-deoxy-1-imino-D-erythrose 4-phosphate: a defining metabolite in the aminoshikimate pathway.
- Guo J, Frost JW[PMID: 11804477]J Am Chem Soc. 124 (2002) 528-9
- Expression and purification of the rifamycin amide synthase, RifF, an enzyme homologous to the prokaryotic arylamine N-acetyltransferases.
- Pompeo F, Mushtaq A, Sim E[PMID: 11812235]Protein Expr Purif. 24 (2002) 138-51
- The loading and initial elongation modules of rifamycin synthetase collaborate to produce mixed aryl ketide products.
- Admiraal SJ, Khosla C, Walsh CT[PMID: 11955082]Biochemistry. 41 (2002) 5313-24
- Kanosamine biosynthesis: a likely source of the aminoshikimate pathway's nitrogen atom.
- Guo J, Frost JW[PMID: 12207504]J Am Chem Soc. 124 (2002) 10642-3
- Characterization of the early stage aminoshikimate pathway in the formation of 3-amino-5-hydroxybenzoic acid: the RifN protein specifically converts kanosamine into kanosamine 6-phosphate.
- Arakawa K, Muller R, Mahmud T, Yu TW, Floss HG[PMID: 12207505]J Am Chem Soc. 124 (2002) 10644-5
- Isolation and characterization of 27-O-demethylrifamycin SV methyltransferase provides new insights into the post-PKS modification steps during the biosynthesis of the antitubercular drug rifamycin B by Amycolatopsis mediterranei S699.
- Xu J, Mahmud T, Floss HG[PMID: 12623077]Arch Biochem Biophys. 411 (2003) 277-88
- A homologue of the Mycobacterium tuberculosis PapA5 protein, rif-orf20, is an acetyltransferase involved in the biosynthesis of antitubercular drug rifamycin B by Amycolatopsis mediterranei S699.
- Xiong Y, Wu X, Mahmud T[PMID: 15791687]Chembiochem. 6 (2005) 834-7
- Identification of tailoring genes involved in the modification of the polyketide backbone of rifamycin B by Amycolatopsis mediterranei S699.
- Xu J, Wan E, Kim CJ, Floss HG, Mahmud T[PMID: 16079331]Microbiology. 151 (2005) 2515-28
- RifP; a membrane protein involved in rifamycin export in Amycolatopsis mediterranei.
- Absalon AE, Fernandez FJ, Olivares PX, Barrios-Gonzalez J, Campos C, Mejia A[PMID: 17351715]Biotechnol Lett. 29 (2007) 951-8
- Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway.
- Claxton HB, Akey DL, Silver MK, Admiraal SJ, Smith JL[PMID: 19103602]J Biol Chem. 284 (2009) 5021-9
- The biosynthesis of 3-amino-5-hydroxybenzoic acid (AHBA), the precursor of mC7N units in ansamycin and mitomycin antibiotics: a review.
- Floss HG, Yu TW, Arakawa K[PMID: 21081954]J Antibiot (Tokyo). 64 (2011) 35-44
- Stereochemistry of reductions catalyzed by methyl-epimerizing ketoreductase domains of polyketide synthases.
- You YO, Khosla C, Cane DE[PMID: 23659177]J Am Chem Soc. 135 (2013) 7406-9
- Coupled methyl group epimerization and reduction by polyketide synthase ketoreductase domains. Ketoreductase-catalyzed equilibrium isotope exchange.
- Garg A, Khosla C, Cane DE[PMID: 24161343]J Am Chem Soc. 135 (2013) 16324-7
- Structure and stereospecificity of the dehydratase domain from the terminal module of the rifamycin polyketide synthase.
- Gay D, You YO, Keatinge-Clay A, Cane DE[PMID: 24274103]Biochemistry. 52 (2013) 8916-28
- Modification of rifamycin polyketide backbone leads to improved drug activity against rifampicin-resistant Mycobacterium tuberculosis.
- Nigam A, Almabruk KH, Saxena A, Yang J, Mukherjee U, Kaur H, Kohli P, Kumari R, Singh P, Zakharov LN, Singh Y, Mahmud T, Lal R[PMID: 24923585]J Biol Chem. 289 (2014) 21142-52
- Substrate structure-activity relationships guide rational engineering of modular polyketide synthase ketoreductases.
- Bailey CB, Pasman ME, Keatinge-Clay AT[PMID: 26568113]Chem Commun (Camb). 52 (2016) 792-5
- Protein-Protein Interactions, Not Substrate Recognition, Dominate the Turnover of Chimeric Assembly Line Polyketide Synthases.
- Klaus M, Ostrowski MP, Austerjost J, Robbins T, Lowry B, Cane DE, Khosla C[PMID: 27246853]J Biol Chem. 291 (2016) 16404-15
 Data download
Data download
 History
History
- 2016-12-27[Update]
- 2014-06-25[Update]
- 2013-12-27[Update]
- 2013-09-04[Update]
- 2013-05-28[Update]
- 2012-10-02[Update]
- 2012-03-28[Release]






