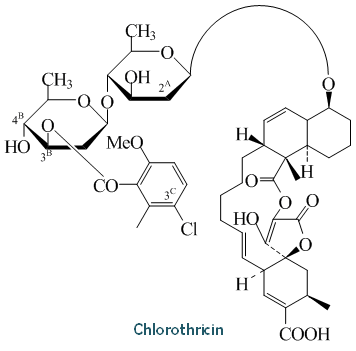
Chlorothricin : Cluster information
 Compound
Compound
| Entry name | Chlorothricin |
|---|---|
| Moiety description | A: aglycone B: 5-chloro-6-methyl-O-methylsalicylic acid moiety |
| PKS Type | A: TypeI modular B: TypeI iterative |
| Starter Unit | A: malonyl-CoA B: acetyl-CoA |
| Chain Length | A: 12 B: 4 |
| Sugar Unit | D-olivose |
| Classification | Tetronic Acid Derivative |
| Activity | Antibacterial |
| Composition | C50H63ClO16 |
 Original source
Original source
| Organism | Streptomyces antibioticus |
|---|---|
| Strain | DSM 40725 |
| Contig | DQ116941 |
 PKS/NRPS Module
PKS/NRPS Module
| A | Chth_00260 chlA1 | 0 |  | malonyl-CoA |
|---|---|---|---|---|
| 1 |  | methylmalonyl-CoA | ||
| 2 |  | malonyl-CoA | ||
| Chth_00270 chlA2 | 3 |  | malonyl-CoA | |
| 4 |  | malonyl-CoA | ||
| Chth_00290 chlA3 | 5 |  | malonyl-CoA | |
| 6 |  | malonyl-CoA | ||
| 7 |  | malonyl-CoA | ||
| Chth_00300 chlA4 | 8 |  | malonyl-CoA | |
| Chth_00310 chlA5 | 9 |  | malonyl-CoA | |
| 10 |  | methylmalonyl-CoA | ||
| Chth_00320 chlA6 | 11 |  | malonyl-CoA | |
| B | Chth_00060 chlB1 | 1 |  | acetyl-CoA malonyl-CoA |
 Reference
Reference
- Cloning and characterization of a bacterial iterative type I polyketide synthase gene encoding the 6-methylsalicyclic acid synthase.
- Shao L, Qu XD, Jia XY, Zhao QF, Tian ZH, Wang M, Tang GL, Liu W[PMID: 16677607]Biochem Biophys Res Commun. 345 (2006) 133-9
- Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis.
- Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, Tang J, Tang GL, Liu W[PMID: 16793515]Chem Biol. 13 (2006) 575-85
- Cloning and characterization of the tetrocarcin A gene cluster from Micromonospora chalcea NRRL 11289 reveals a highly conserved strategy for tetronate biosynthesis in spirotetronate antibiotics.
- Fang J, Zhang Y, Huang L, Jia X, Zhang Q, Zhang X, Tang G, Liu W[PMID: 18586939]J Bacteriol. 190 (2008) 6014-25
- Dissection of two acyl-transfer reactions centered on acyl-S-carrier protein intermediates for incorporating 5-chloro-6-methyl-O-methylsalicyclic acid into chlorothricin.
- He QL, Jia XY, Tang MC, Tian ZH, Tang GL, Liu W[PMID: 19266533]Chembiochem. 10 (2009) 813-9
- Insights into bacterial 6-methylsalicylic acid synthase and its engineering to orsellinic acid synthase for spirotetronate generation.
- Ding W, Lei C, He Q, Zhang Q, Bi Y, Liu W[PMID: 20534347]Chem Biol. 17 (2010) 495-503
- Coordinative Modulation of Chlorothricin Biosynthesis by Binding of the Glycosylated Intermediates and End Product to a Responsive Regulator ChlF1.
- Li Y, Li J, Tian Z, Xu Y, Zhang J, Liu W, Tan H[PMID: 26750095]J Biol Chem. 291 (2016) 5406-17
- Insights into 6-Methylsalicylic Acid Bio-assembly by Using Chemical Probes.
- Parascandolo JS, Havemann J, Potter HK, Huang F, Riva E, Connolly J, Wilkening I, Song L, Leadlay PF, Tosin M[PMID: 26833898]Angew Chem Int Ed Engl. 55 (2016) 3463-7
 Data download
Data download
 History
History
- 2016-12-27[Update]
- 2012-10-02[Update]
- 2012-08-03[Update]
- 2012-04-26[Release]






