Virginiamycin : Cluster information
 Compound
Compound
| Entry name | Virginiamycin |
|---|---|
| PKS Type | Virginiamycin M: PKS-NRPS hybrid Virginiamycin M: Trans-AT Virginiamycin S: NRPS |
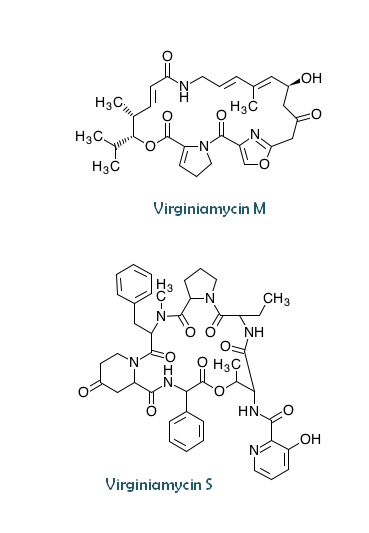
| Starter Unit | Virginiamycin M: isobutyryl-CoA Virginiamycin S: 3-hydroxypicolinic acid |
| Chain Length | Virginiamycin M: 7 |
| Sugar Unit | none |
| Classification | Streptogramin |
| Activity | Antibacterial |
| Composition | Virginiamycin M: C28H35N3O7 Virginiamycin S: C43H49N7O10 |
 Original source
Original source
| Organism | Streptomyces virginiae |
|---|---|
| Strain | MAFF 10-06014 |
| Contig | AB621357 AB283030 AB253363 AB253364 AB035547 AB035548 AB001608 D32251 AB001609 AB019519 AB046994 AB072568 |
 PKS/NRPS Module
PKS/NRPS Module
| M | Virg_00250 virA | 1 |  | |
|---|---|---|---|---|
| 2 | 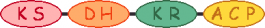 | |||
| 3 |  | glycine | ||
| 4 |  | |||
| 5 |  | |||
| Virg_00200 virF | 6 |  | ||
| Virg_00190 virG | 6 | 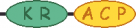 | ||
| 7 | 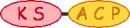 | |||
| Virg_00180 virH | 8 |  | serine | |
| 9 | 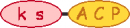 | |||
| Virg_00170 virI | 10 |  | malonyl-CoA not conserved HAFHS(H->G,A->P) | |
| S | Virg_00390 visB | 0 |  | |
| Virg_00090 visE | 1 |  | L-threonine | |
| 2 | 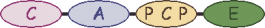 | L-aminobutyric acid | ||
| Virg_00080 visF | 3 |  | proline | |
| 4 | 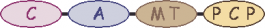 | phenylalanine | ||
| 5 |  | L-pipecolate | ||
| 6 | 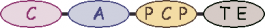 | L-phenylglycine |
 Reference
Reference
- Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae.
- Kinoshita H, Ipposhi H, Okamoto S, Nakano H, Nihira T, Yamada Y[PMID: 9371444]J Bacteriol. 179 (1997) 6986-93
- Identification and in vivo functional analysis of a virginiamycin S resistance gene (varS) from Streptomyces virginiae.
- Lee CK, Kamitani Y, Nihira T, Yamada Y[PMID: 10322037]J Bacteriol. 181 (1999) 3293-7
- Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance.
- Kawachi R, Akashi T, Kamitani Y, Sy A, Wangchaisoonthorn U, Nihira T, Yamada Y[PMID: 10792718]Mol Microbiol. 36 (2000) 302-13
- Identification by gene deletion analysis of a regulator, VmsR, that controls virginiamycin biosynthesis in Streptomyces virginiae.
- Kawachi R, Wangchaisoonthorn U, Nihira T, Yamada Y[PMID: 11029453]J Bacteriol. 182 (2000) 6259-63
- Identification of the varR gene as a transcriptional regulator of virginiamycin S resistance in Streptomyces virginiae.
- Namwat W, Lee CK, Kinoshita H, Yamada Y, Nihira T[PMID: 11222601]J Bacteriol. 183 (2001) 2025-31
- Characterization of virginiamycin S biosynthetic genes from Streptomyces virginiae.
- Namwat W, Kamioka Y, Kinoshita H, Yamada Y, Nihira T[PMID: 11943483]Gene. 286 (2002) 283-90
- Identification by heterologous expression and gene disruption of VisA as L-lysine 2-aminotransferase essential for virginiamycin S biosynthesis in Streptomyces virginiae.
- Namwat W, Kinoshita H, Nihira T[PMID: 12169606]J Bacteriol. 184 (2002) 4811-8
- barS1, a gene for biosynthesis of a gamma-butyrolactone autoregulator, a microbial signaling molecule eliciting antibiotic production in Streptomyces species.
- Shikura N, Yamamura J, Nihira T[PMID: 12193632]J Bacteriol. 184 (2002) 5151-7
- Identification by gene deletion analysis of barB as a negative regulator controlling an early process of virginiamycin biosynthesis in Streptomyces virginiae.
- Matsuno K, Yamada Y, Lee CK, Nihira T[PMID: 14647980]Arch Microbiol. 181 (2004) 52-9
- Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae.
- Pulsawat N, Kitani S, Nihira T[PMID: 17350183]Gene. 393 (2007) 31-42
- Identification of the bkdAB gene cluster, a plausible source of the starter-unit for virginiamycin M production in Streptomyces virginiae.
- Pulsawat N, Kitani S, Kinoshita H, Lee CK, Nihira T[PMID: 17375285]Arch Microbiol. 187 (2007) 459-66
- Identification by gene deletion analysis of barS2, a gene involved in the biosynthesis of gamma-butyrolactone autoregulator in Streptomyces virginiae.
- Lee YJ, Kitani S, Kinoshita H, Nihira T[PMID: 18034227]Arch Microbiol. 189 (2008) 367-74
- Hierarchical control of virginiamycin production in Streptomyces virginiae by three pathway-specific regulators: VmsS, VmsT and VmsR.
- Pulsawat N, Kitani S, Fukushima E, Nihira T[PMID: 19332826]Microbiology. 155 (2009) 1250-9
- Null mutation analysis of an afsA-family gene, barX, that is involved in biosynthesis of the {gamma}-butyrolactone autoregulator in Streptomyces virginiae.
- Lee YJ, Kitani S, Nihira T[PMID: 19778967]Microbiology. 156 (2010) 206-10
- Crystallization and preliminary crystallographic studies of the butyrolactone autoregulator receptor protein (BarA) from Streptomyces virginiae.
- Yoon YH, Kawai F, Sugiyama K, Park SY, Nihira T, Choi SU, Hwang YI[PMID: 20516594]Acta Crystallogr Sect F Struct Biol Cryst Commun. 66 (2010) 662-4
- VisG is essential for biosynthesis of virginiamycin S, a streptogramin type B antibiotic, as a provider of the nonproteinogenic amino acid phenylglycine.
- Ningsih F, Kitani S, Fukushima E, Nihira T[PMID: 21816878]Microbiology. 157 (2011) 3213-20
- Characterization of Intersubunit Communication in the Virginiamycin trans-Acyl Transferase Polyketide Synthase.
- Dorival J, Annaval T, Risser F, Collin S, Roblin P, Jacob C, Gruez A, Chagot B, Weissman KJ[PMID: 26982529]J Am Chem Soc. 138 (2016) 4155-67
 Data download
Data download
 History
History
- 2016-12-27[Update]
- 2016-01-27[Update]
- 2013-05-28[Update]
- 2013-01-16[Update]
- 2012-10-02[Update]
- 2012-08-03[Release]







