Nysta_00130 : CDS information
 Location
Location
| Organism | Streptomyces noursei |
|---|---|
| Strain | ATCC 11455 (=NBRC 15452) |
| Entry name | Nystatin |
| Contig | AF263912 |
| Start / Stop / Direction | 62,659 / 66,759 / + [in whole cluster] 62,659 / 66,759 / + [in contig] |
| Location | 62659..66759 [in whole cluster] 62659..66759 [in contig] |
| Type | CDS |
| Length | 4,101 bp (1,366 aa) |
 Annotation
Annotation
| Category | 1.1 PKS |
|---|---|
| Product | polyketide synthase |
| Product (GenBank) | NysA |
| Gene | |
| Gene (GenBank) | nysA |
| EC number | |
| Keyword | |
| Note | |
| Note (GenBank) |
|
| Reference |
|
 PKS/NRPS Module
PKS/NRPS Module
| 0 | 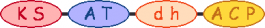 | acetyl-CoA |
 Sequence
Sequence
>polyketide synthase [NysA] MTIGADEDPVVVVGMACRYPGGVAGPEDLWELVRTGRDATTAFPDDRGWDLAALAGDGPG RSATREGGFLTGAADFDAAFFGMSPREAVSTDPQQRLVLETAWEALERAGIDPHSLRGSR TGVFVGASGQDYAAVTHASPDDLDGHALTGLAPGVASGRLAYVLGLEGPAVTVDTTSSSS LVALHWAVRALRAGECSTALAGGVTVMSTPAAFVGHTRQGGLAPDGRCKPFSDDADGTAW AEGVGIVVLEHLSTARAAGNPVLAVLRGSAVNQDGASDGLTAPSGPAQERVIRAALADAR LAPADIDLVEAHGTGTRLGDPVEARALLAAYGQDRDPDRPLRLGSLKSTLGHAQAAAGIG GVIKTVLTLRHGLMPRIRHLATPTRQVDWSQGAVAPLTDHTPWPPADRPRRAGVSSFGIS GTNAHVILEEAPPADVPVTRPGTLRPSTVPWPVSAATPEALDAQLARLRAHLRTHSDLDP LDVGYSLATGRAALRHRAVLLPPADGTAADAVEHARGAAHQRRTAVLFSGQGSQRPGMGR ELAARFPVFADALDDALRALDRHLDGPVREVMWGTDAALLDRTGWTQPALFAVEVALHRL VASLGVTPDFVGGHSVGEIAAAHVAGVLSLEDACRLVAARATLMQALPAGGAMAALEATE DEVAPLLGAHLALAAVNGPTAVVVAGAEDAVRQLTARFADRGRRTSRLAVSHAFHSPLME PMLDAFRDVVSRLTFHQPSIPLVSNLTGELAGSEITSAEYWVRHVRDTVRFADGITALAK AGADVLIELGPGGVLSAMARDTLGPDSTTDVVPALSKGRPEETAFAGALGRLHTLGVPVD WPAFYAGTGARRVELPTYAFQHVRHWPTPPRPNGAGPGALGHPLLGSAVELADGGGTVCS GALSLRTHPWLADHTVAGRVVLPATALLELAVRAGDEAGCDVLHELHLTTPPALPDDAAL HVQVHVGPADTTGRRAVTVHTRPDHHPAGDWTRCATGTLGSTPPSAAEAATGGTPAAWPP ADAEPLDLADHYERLADRGFDYGPTFRGLRAAWRRGAEIFADVECPPGTADDAPDHGLHP ALLDAARHAAMAVDGTVPVAWHGVRLHAVGATALRVRIRPTTTGTLTLTAVDVHGAPVVT VEALTARPLTDEERAAPRTPRQARGETPADARPARPAAARPGPAGEPLPDTTGSHPTAGH LAALPPAARERQLLDLVRTQAAAVLGHPGPEAVGTRSVFKELGFDSLAGVELADRLTART GLRLPATLVFNFPTPERAAHRLGELLAATAPLDPGAYGEELTRFEAIVTNLPQDGPERRA VADRLDAIVSALRQNSPAEVPSSDEDIDTVSVDRLLDIIDEEFETT
>polyketide synthase [NysA] ATGACGATCGGAGCCGACGAGGACCCGGTGGTGGTCGTCGGAATGGCCTGCCGTTATCCG GGTGGGGTCGCCGGCCCGGAGGACCTGTGGGAACTGGTCCGCACCGGCCGCGACGCGACC ACCGCCTTCCCGGACGACCGCGGCTGGGACCTGGCCGCACTGGCCGGCGACGGACCCGGC CGCAGCGCGACCCGCGAGGGCGGATTCCTCACCGGCGCCGCCGACTTCGACGCCGCCTTC TTCGGCATGTCGCCCCGCGAGGCCGTCTCCACCGACCCGCAACAGCGCCTCGTCCTGGAG ACCGCCTGGGAAGCCCTGGAGCGCGCCGGCATCGACCCGCACTCCCTGCGCGGCAGCCGC ACCGGGGTCTTCGTCGGCGCCAGCGGCCAGGACTACGCCGCCGTCACCCACGCCTCGCCC GACGACCTGGACGGACACGCCCTCACCGGCCTGGCCCCCGGCGTCGCCTCCGGTCGCCTG GCGTACGTCCTGGGCCTCGAAGGCCCCGCCGTCACCGTCGACACCACGTCCTCCTCGTCG CTGGTCGCGCTGCACTGGGCGGTCCGCGCCCTGCGCGCGGGGGAGTGCAGCACCGCCCTG GCCGGCGGCGTCACGGTGATGTCCACCCCGGCCGCCTTCGTCGGCCACACCCGACAGGGC GGCCTCGCGCCCGACGGCCGCTGCAAGCCGTTCTCCGACGACGCCGACGGCACCGCCTGG GCGGAGGGCGTCGGCATCGTCGTCCTGGAGCACCTGTCCACCGCCCGCGCCGCCGGCAAC CCCGTCCTCGCCGTGCTGCGCGGCTCGGCCGTCAACCAGGACGGCGCCTCCGACGGCCTC ACCGCACCCAGCGGTCCCGCCCAGGAACGCGTCATCCGCGCCGCCCTCGCCGACGCCCGA CTCGCCCCCGCCGACATCGATCTCGTCGAGGCGCACGGCACCGGCACCCGGCTCGGCGAC CCCGTCGAGGCCCGGGCGCTGCTCGCCGCCTACGGCCAGGACCGGGACCCGGACCGACCG CTGCGCCTCGGTTCCCTGAAGTCCACCCTCGGCCACGCACAGGCCGCCGCCGGCATCGGC GGAGTGATCAAGACCGTCCTGACCCTGCGGCACGGCCTGATGCCGCGCATCCGGCACCTG GCCACCCCCACCCGCCAAGTCGACTGGTCCCAGGGCGCCGTGGCCCCCCTCACCGACCAC ACGCCCTGGCCACCGGCCGACCGACCGCGCCGCGCCGGCGTCTCCTCCTTCGGCATCAGC GGCACCAACGCCCATGTGATCCTCGAAGAGGCGCCGCCCGCCGACGTCCCCGTCACCCGG CCCGGCACCCTCCGCCCCAGCACCGTCCCCTGGCCGGTCTCCGCCGCCACGCCCGAAGCC CTCGACGCCCAACTCGCCCGGCTCCGCGCCCACCTGCGCACCCACTCGGACCTGGACCCG CTGGACGTCGGCTACTCCCTGGCCACCGGCCGCGCCGCGCTCCGCCACCGGGCGGTCCTC CTGCCGCCCGCCGACGGCACCGCCGCGGACGCCGTCGAGCACGCCCGCGGTGCGGCCCAC CAGCGCCGCACCGCCGTCCTCTTCTCCGGCCAGGGCAGCCAGCGCCCGGGCATGGGCCGC GAACTCGCCGCCCGCTTCCCGGTGTTCGCCGACGCACTGGACGACGCGCTGCGCGCCCTG GACCGGCACCTGGACGGCCCGGTGCGCGAGGTGATGTGGGGCACCGACGCCGCGCTCCTG GACCGGACCGGCTGGACCCAGCCCGCCCTGTTCGCCGTCGAGGTCGCCCTCCACCGCCTG GTCGCGTCCCTCGGCGTCACCCCCGACTTCGTCGGCGGCCACTCCGTCGGCGAGATCGCC GCCGCCCACGTCGCCGGCGTCCTGAGCCTGGAGGACGCCTGCCGCCTGGTGGCCGCCCGC GCCACGCTGATGCAGGCGCTCCCGGCCGGCGGCGCGATGGCCGCGCTGGAGGCCACCGAG GACGAAGTGGCCCCGCTGCTCGGCGCACACCTCGCGCTGGCCGCCGTCAACGGCCCCACC GCGGTCGTCGTCGCCGGAGCCGAGGACGCCGTGCGGCAACTGACCGCCCGCTTCGCCGAC CGCGGCCGGCGCACCAGCCGGCTGGCCGTCTCGCACGCCTTCCACTCGCCGCTGATGGAG CCCATGCTCGACGCCTTCCGGGACGTCGTGAGCCGACTGACCTTCCACCAGCCGTCGATC CCGCTGGTCTCCAACCTCACCGGTGAACTCGCCGGCAGTGAGATCACCAGCGCCGAGTAC TGGGTCCGGCACGTCCGCGACACCGTCCGCTTCGCCGACGGCATCACCGCACTGGCCAAG GCCGGCGCCGACGTCCTGATCGAACTCGGCCCCGGCGGCGTGCTGTCCGCGATGGCCCGC GACACCCTCGGCCCCGACAGCACCACCGACGTCGTCCCCGCCCTGAGCAAGGGACGGCCC GAGGAGACCGCCTTCGCCGGCGCCCTCGGCCGCCTGCACACCCTCGGCGTCCCCGTCGAC TGGCCCGCCTTCTACGCCGGCACCGGCGCCCGCCGCGTCGAACTGCCCACCTACGCCTTC CAGCACGTGCGCCACTGGCCCACCCCGCCCCGCCCGAACGGCGCCGGGCCCGGCGCCCTC GGCCACCCCCTGCTCGGCTCCGCCGTCGAACTCGCCGACGGCGGCGGCACCGTCTGCTCC GGCGCCCTCTCCCTCCGCACCCACCCCTGGCTCGCCGACCACACCGTCGCCGGGCGGGTC GTGCTGCCGGCCACCGCGCTGCTGGAACTCGCCGTGCGCGCCGGCGACGAGGCGGGCTGC GACGTCCTGCACGAACTCCACCTCACCACCCCGCCGGCCCTGCCCGACGACGCCGCCCTG CACGTCCAGGTGCACGTCGGCCCCGCCGACACCACCGGGCGCCGCGCCGTCACCGTCCAC ACCCGCCCCGACCACCACCCGGCCGGCGACTGGACCCGATGCGCCACCGGCACCCTCGGC AGCACCCCGCCGTCCGCAGCCGAAGCCGCCACGGGCGGCACCCCGGCCGCCTGGCCGCCG GCCGACGCCGAACCCCTCGACCTCGCCGACCACTACGAACGGCTCGCCGACCGCGGCTTC GACTACGGCCCGACCTTCCGCGGCCTGCGGGCCGCCTGGCGACGCGGCGCGGAGATCTTC GCGGACGTGGAATGCCCGCCCGGCACCGCCGACGACGCCCCCGACCACGGACTGCACCCC GCCCTGCTCGACGCGGCCCGGCACGCCGCCATGGCGGTGGACGGCACCGTGCCCGTCGCC TGGCACGGCGTCCGGCTGCACGCCGTCGGCGCCACCGCGCTGCGGGTCCGCATCCGCCCC ACCACGACCGGCACGCTGACCCTCACCGCGGTCGACGTGCACGGCGCGCCGGTCGTCACC GTCGAGGCCCTCACCGCCCGCCCGCTGACCGACGAGGAACGCGCCGCCCCGCGGACGCCG CGGCAGGCCCGCGGCGAGACGCCCGCCGACGCCCGCCCGGCCCGGCCCGCGGCGGCCCGC CCCGGCCCGGCCGGCGAACCCCTCCCGGACACCACCGGGTCCCACCCCACCGCCGGCCAC CTCGCCGCGCTGCCGCCGGCCGCCCGGGAGCGCCAGCTGCTGGACCTGGTGCGCACCCAG GCCGCCGCCGTCCTGGGCCACCCCGGCCCCGAGGCCGTCGGCACCCGCAGCGTCTTCAAG GAGCTGGGCTTCGACTCGTTGGCCGGCGTCGAACTCGCCGACCGGCTCACCGCCCGCACC GGACTGCGCCTGCCGGCCACCCTCGTCTTCAACTTCCCCACCCCCGAACGTGCTGCCCAC CGCCTCGGGGAACTCCTCGCCGCAACCGCCCCCCTCGACCCCGGGGCGTACGGAGAGGAA CTCACCAGGTTCGAGGCGATCGTGACGAACCTGCCGCAGGACGGCCCCGAACGCCGGGCC GTCGCGGACCGGTTGGACGCCATCGTCTCCGCACTCCGCCAGAACTCGCCTGCAGAGGTG CCCTCCTCGGACGAGGACATCGACACGGTGTCGGTCGACAGACTGCTCGACATCATCGAT GAAGAGTTCGAAACCACATAG
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR001227 Acyl transferase domain (Domain)
IPR006162 Phosphopantetheine attachment site (PTM)
IPR009081 Acyl carrier protein-like (Domain)
IPR014030 Beta-ketoacyl synthase, N-terminal (Domain)
IPR014031 Beta-ketoacyl synthase, C-terminal (Domain)
IPR014043 Acyl transferase (Domain)
IPR016035 Acyl transferase/acyl hydrolase/lysophospholipase (Domain)
IPR016036 Malonyl-CoA ACP transacylase, ACP-binding (Domain)
IPR016038 Thiolase-like, subgroup (Domain)
IPR016039 Thiolase-like (Domain)
IPR020801 Polyketide synthase, acyl transferase domain (Domain)
IPR020806 Polyketide synthase, phosphopantetheine-binding domain (Domain)
IPR020807 Polyketide synthase, dehydratase domain (Domain)
IPR020841 Polyketide synthase, beta-ketoacyl synthase domain (Domain)
|
| SignalP | |
| TMHMM | No significant hit |


 Nysta_00120
Nysta_00120


