Virg_00190 : CDS information
 Location
Location
| Organism | Streptomyces virginiae |
|---|---|
| Strain | MAFF 10-06014 |
| Entry name | Virginiamycin |
| Contig | AB283030 |
| Start / Stop / Direction | 58,367 / 54,744 / - [in whole cluster] 29,991 / 26,368 / - [in contig] |
| Location | complement(54744..58367) [in whole cluster] complement(26368..29991) [in contig] |
| Type | CDS |
| Length | 3,624 bp (1,207 aa) |
 Annotation
Annotation
| Category | 1.1 PKS |
|---|---|
| Product | polyketide synthase |
| Product (GenBank) | hybrid non ribosomal peptide synthetase-polyketide synthase |
| Gene | |
| Gene (GenBank) | virG |
| EC number | |
| Keyword |
|
| Note | |
| Note (GenBank) | |
| Reference |
|
 PKS/NRPS Module
PKS/NRPS Module
| M | 6 | 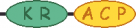 |
|---|---|---|
| 7 | 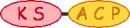 |
 Sequence
Sequence
>polyketide synthase [hybrid non ribosomal peptide synthetase-polyketide synthase] MLTADGTPDARRLLAEIAAPAAEHGHLADLRSARRLRRVLVPVPLPGTRPFVRQDGVYLV SGGAGGIGLALARHLTDRPGTRVVLCGRTAWDDLAEATRSAVSGSDRLRYARADVTDPEA AAALVAEVVRTEGALHGVFHAAGVVRDGYLVRKDPRDVADVLAPKILGARALDEATAALP LDAFVLFSSVAAVTGNLGQSDYAFANGFLDGFATRRAGQVARGERHGRTLSVQWPLWDVP GMSIPEPVLEVVAQHTGMAPLPAAVGLAALERLLAADGPEVVSLFHGDAATWRAHLAALH LERPAAAGQVTAAAPAAPAPSDTPVVASADPAAADDGRRAELADRVSRTVADTIGRPAGS IGGHTSLESMGLDSVMIRALASRLSAEVAPVGPEMLFGLRDLDELVDHLVAARPVPVAEP ATPAVPPAPAATAATVFAPVAPADPAVVPAAPAALPVPAAAPASASRTLPAPSADDRFAI IGISGRYPQAPDLDAFWQNLLNGKDTASDLPTDRWPDAAGVNARGHFLEGVDAFDPTFFG LSAHDGTLLDPQERLFLEVAWEALEDAGYTGSRLHDLVAPDGERRSVGVFAGITSSDYKL LGAERWAAGHREMPSGHYWSLPNRLSYLLDLRGPSQPVDTACSSSLVALHLALDALRRGE CAAALVGGVNLYVHPSRFRMLRRSGFLAEDGLCRSFGAGGAGFGPGEGGGAVVVKPLAVA LADGDTVHAVVRGSAVAHGGRTNGFTAPSPWAQARVLRAALRNSGTDPETVNVIEAHGTG TELGDPVELAGLQDAYGSGRVPCSLGSVKSAVGHGESAAGIAALTKVVLQLRHGELVPTL HADPVNPGLRLEDTRFVLQHTPGRWERLTGADGSPLPRRAGISSFGAGGVNAHVIVEEYL PEPHGRAAAAEPGRPELVLLSAPTREHLAATADRLARRLEGPEAPADLRAVAYSLRTGRS AMDCRLAVVATDTAGLAAALGAFARSADAGEGESAVRYADLRDGRRAHRDLDTVPETTAF LADLWRNRRLHQLGELWLSGLDIERAAPREPGTRLVPLPTSAFLRRRLWITDPVTADTTP TPDPLPTPDPLPVPLSAPAPAPAPAPQPAPTTTPAPAAAPAAAPARSGEAVTEQLSRLVA GFVPDAAGSVDPDRTLLEHGIDSINLMNLRFEITERFGRTLPLQLLSESTVPVLAAHLSA DRAHDRA
>polyketide synthase [hybrid non ribosomal peptide synthetase-polyketide synthase] GTGCTGACCGCGGACGGCACGCCGGACGCGAGGCGGTTGCTCGCCGAGATCGCCGCGCCG GCCGCCGAGCACGGGCACCTGGCCGACCTCCGCTCCGCCCGGCGGCTGCGCCGGGTGCTG GTACCGGTTCCGCTGCCCGGCACCCGCCCCTTCGTGCGCCAGGACGGCGTGTACCTGGTC AGCGGCGGTGCGGGCGGCATCGGCCTGGCACTGGCCCGGCACCTGACGGACCGTCCGGGC ACCCGCGTGGTGCTCTGCGGACGGACCGCGTGGGACGACCTCGCCGAAGCGACCCGCAGC GCCGTCTCCGGCAGCGACCGCCTGCGCTACGCCAGGGCCGACGTCACCGACCCCGAGGCC GCCGCCGCGCTCGTGGCCGAGGTCGTCCGGACCGAGGGCGCCCTGCACGGCGTCTTCCAC GCCGCCGGCGTGGTGCGCGACGGCTACCTGGTGCGCAAGGACCCCCGGGACGTGGCCGAC GTGCTGGCGCCCAAGATCCTCGGTGCGCGGGCGCTGGACGAGGCGACCGCGGCCCTCCCG CTGGACGCGTTCGTGCTGTTCTCCTCGGTGGCCGCGGTGACCGGCAACCTCGGCCAGAGC GACTACGCGTTCGCCAACGGCTTCCTCGACGGCTTCGCCACCCGCCGTGCCGGGCAGGTG GCGCGCGGTGAGCGGCACGGCAGGACGCTGTCCGTGCAGTGGCCGCTCTGGGACGTCCCC GGCATGAGCATTCCCGAGCCGGTCCTGGAAGTGGTGGCCCAGCACACCGGCATGGCGCCG CTGCCCGCAGCCGTCGGGCTCGCGGCTCTGGAACGCCTGCTGGCCGCCGACGGCCCCGAG GTCGTCTCGCTCTTTCACGGCGACGCGGCCACGTGGCGGGCCCACCTGGCCGCCCTCCAC CTGGAGCGGCCGGCCGCGGCCGGCCAGGTGACGGCCGCCGCTCCCGCCGCTCCCGCCCCT TCCGACACCCCCGTCGTCGCGTCGGCCGATCCTGCTGCGGCGGATGACGGCCGGCGCGCC GAACTGGCCGACCGGGTCAGCCGGACGGTGGCCGACACCATCGGCCGCCCCGCGGGCAGC ATCGGCGGTCACACCTCGCTCGAGTCGATGGGGCTGGACTCCGTCATGATCCGCGCCCTC GCCTCCCGGCTCAGCGCCGAAGTGGCACCGGTCGGGCCGGAGATGCTCTTCGGCCTGCGC GACCTGGACGAGCTGGTCGACCACCTGGTCGCCGCCCGGCCGGTCCCCGTAGCGGAACCG GCCACACCGGCGGTCCCGCCGGCACCCGCCGCCACGGCGGCCACGGTCTTCGCACCGGTG GCTCCGGCGGACCCGGCCGTTGTCCCGGCCGCCCCGGCCGCCCTGCCCGTCCCCGCCGCC GCGCCGGCATCCGCCTCCCGTACGCTGCCGGCGCCGTCCGCCGATGACCGCTTCGCGATC ATCGGCATCAGCGGCCGCTACCCGCAGGCGCCCGACCTGGACGCCTTCTGGCAGAACCTG CTGAACGGCAAGGACACCGCCTCCGACCTGCCCACCGACCGCTGGCCGGACGCCGCGGGC GTCAACGCCCGGGGCCACTTCCTCGAAGGCGTGGACGCGTTCGACCCGACGTTCTTCGGA CTGTCCGCCCATGACGGGACCCTGCTCGACCCGCAGGAACGGCTGTTCCTGGAGGTCGCC TGGGAGGCGCTGGAGGATGCCGGATACACCGGCTCGCGTCTGCACGACCTCGTCGCGCCG GACGGCGAGCGCCGCAGCGTCGGCGTCTTCGCCGGGATCACCTCCAGCGACTACAAGCTG CTGGGCGCCGAGCGCTGGGCAGCCGGACACCGGGAGATGCCCTCCGGGCACTACTGGAGC CTGCCCAACCGGCTGTCCTACCTGCTGGACCTGCGCGGTCCGAGCCAGCCGGTCGACACC GCCTGCTCGTCCTCCCTGGTCGCGCTGCACCTCGCGCTCGACGCGCTCCGCCGCGGCGAG TGCGCGGCCGCTCTGGTCGGCGGGGTCAACCTGTACGTCCACCCCTCCCGGTTCCGGATG CTGCGCCGGTCCGGCTTCCTGGCCGAGGACGGCCTGTGCCGCAGCTTCGGCGCCGGCGGG GCCGGCTTCGGCCCGGGCGAGGGCGGCGGTGCCGTCGTGGTGAAACCGCTCGCCGTCGCC CTGGCCGACGGTGACACCGTGCACGCGGTGGTCCGCGGCAGCGCGGTCGCACACGGCGGG CGGACCAACGGCTTCACCGCGCCGAGCCCGTGGGCGCAGGCACGGGTGCTCCGTGCCGCG CTGCGGAACTCCGGCACCGACCCGGAGACCGTCAACGTGATCGAGGCGCACGGCACCGGG ACGGAACTCGGCGACCCGGTCGAGCTGGCCGGCCTCCAGGACGCGTACGGCTCCGGCCGC GTGCCCTGCTCGCTGGGGTCGGTCAAGTCCGCGGTGGGGCACGGCGAGTCGGCGGCCGGC ATCGCCGCGCTGACCAAGGTGGTCCTCCAACTGCGCCACGGCGAGCTGGTCCCCACCCTG CACGCCGACCCGGTCAACCCGGGCCTGCGCCTGGAGGACACCCGCTTCGTCCTCCAGCAC ACCCCGGGCCGCTGGGAGCGACTGACCGGCGCGGACGGCAGCCCGCTGCCGCGACGGGCG GGCATCAGCTCCTTCGGCGCCGGCGGAGTCAACGCCCACGTCATCGTCGAGGAGTACCTG CCCGAGCCGCACGGCCGCGCGGCCGCCGCCGAGCCGGGCCGCCCGGAACTGGTCCTGCTC TCCGCCCCGACGCGCGAGCACCTGGCGGCCACCGCCGACCGCCTCGCGCGCCGGCTGGAG GGCCCGGAGGCGCCCGCCGACCTGCGGGCGGTGGCGTACAGCCTGCGCACCGGCCGGTCC GCCATGGACTGTCGCCTCGCCGTGGTGGCGACCGACACCGCCGGCCTGGCGGCCGCGCTC GGCGCCTTCGCCCGCTCGGCCGACGCCGGCGAGGGGGAGTCCGCCGTGCGGTACGCCGAC CTGCGGGACGGCCGCCGTGCCCATCGGGACCTGGACACGGTGCCCGAGACGACCGCGTTC CTCGCCGACCTGTGGCGCAACCGGCGCCTGCACCAACTCGGCGAGCTCTGGCTGTCGGGC CTCGACATCGAGCGTGCCGCGCCCCGTGAGCCGGGCACCCGGCTGGTGCCGCTCCCCACG TCGGCGTTCCTGCGGCGCCGGCTCTGGATCACCGACCCGGTCACGGCGGACACCACGCCC ACACCGGACCCTCTGCCCACACCGGACCCCCTGCCCGTACCGCTGTCCGCTCCGGCACCG GCACCGGCCCCCGCCCCCCAGCCCGCGCCGACCACCACCCCGGCCCCGGCAGCCGCGCCC GCCGCAGCGCCTGCCAGATCCGGGGAAGCGGTGACCGAGCAGCTGTCGCGCCTCGTCGCG GGCTTCGTACCCGACGCGGCGGGGTCGGTCGACCCCGACCGCACCCTGCTCGAACACGGC ATCGACTCGATCAACCTGATGAACCTGCGCTTCGAGATCACCGAGCGGTTCGGGCGGACC CTGCCGCTGCAGTTGCTCAGCGAGTCGACCGTCCCCGTGCTCGCCGCCCACCTGTCCGCC GACCGCGCCCACGACCGGGCCTGA
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR009081 Acyl carrier protein-like (Domain)
IPR013968 Polyketide synthase, KR (Domain)
IPR014030 Beta-ketoacyl synthase, N-terminal (Domain)
IPR014031 Beta-ketoacyl synthase, C-terminal (Domain)
IPR016038 Thiolase-like, subgroup (Domain)
IPR016039 Thiolase-like (Domain)
IPR016040 NAD(P)-binding domain (Domain)
IPR018201 Beta-ketoacyl synthase, active site (Active_site)
IPR020842 Polyketide synthase/Fatty acid synthase, KR (Domain)
|
| SignalP | |
| TMHMM | No significant hit |


 Virg_00180
Virg_00180


