Oxzol_00220 : CDS information
 Location
Location
| Organism | Streptomyces albus |
|---|---|
| Strain | JA3453 |
| Entry name | Oxazolomycin |
| Contig | EF552687 |
| Start / Stop / Direction | 85,129 / 87,657 / + [in whole cluster] 85,129 / 87,657 / + [in contig] |
| Location | 85129..87657 [in whole cluster] 85129..87657 [in contig] |
| Type | CDS |
| Length | 2,529 bp (842 aa) |
 Annotation
Annotation
| Category | 1.1 PKS |
|---|---|
| Product | polyketide synthase |
| Product (GenBank) | PKS |
| Gene | |
| Gene (GenBank) | ozmQ |
| EC number | |
| Keyword | |
| Note | |
| Note (GenBank) | |
| Reference |
|
 PKS/NRPS Module
PKS/NRPS Module
| 1 | 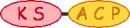 |
 Sequence
Sequence
>polyketide synthase [PKS] MNDQRTEQQAAAAEVRDSDIAVIGLACRFPGAATPDTFWKVLSEGRETLTHFSDEELRAA GVAEPLLADDRYVKAGQVLVDADKFDAGLFGITRDEAELIDPQQRQFLECAYEALERAGY DPQRGEQRIGVYAGVGLNTYLLHNLGERYRTASSVDRYRMMITNDKDFVATRTAYKLNLC GPSVSTNTACSTSLVAVHLACLSLLSGDCTMALAGAAHIQADQGEGYLHHEGMIFSPDGH CRAFDAKAQGTVIGNGVGAVVLKRLSDALADGDTVHAVIKGTAVNNDGSDKTGYTAPSVQ GQAAVVAEAQEIADVGPETVSYVEAHGTATPLGDPIEVAALNQAFNREGAALAPGSCALG SVKTNVGHLDTAAGMAGLIKTILMLRHRTLVPSLCFEAPNPEIDFAAGPFYVGTETKEWP AGPTPRRAGVSSFGIGGTNAHVIVEEPPAVGPAPDAAAAGPRLLVLSANTPAALDTATAD LARALRKDRDLDLSAVAQTLALGRRVLPYRRALVATGVRDAALALALGDAGRVMTAGPAD ERPVVELVTGGGTPEHAAALYEEAAAFREHFDRCAAELGTPAAELLRGHGPDAAFAVQYA TARALAGWGSTAPVVAADRTELPDAALRLLDGIGAQHTAGAGRPGVALLPAASAPVGTAF LLGLIGRLWTAGDTVDWTVFHQGEPVRRVPLPTYPFERVRHWAEPHGGAAPAAGPAALTL RDRFQGVPDAEKSALLTGFIRQEIAAVMGASDPRTVDLDTNLFDMGLDSLILIEVIAKLS DELAHPVRSSAFVEYPTVRSFVADLGEELGIVPGTPEAAPAASGGRVSRRAQRAAARRPG GA
>polyketide synthase [PKS] ATGAACGACCAGCGAACAGAGCAGCAGGCAGCCGCGGCCGAGGTCCGGGACTCCGACATC GCCGTCATCGGGCTCGCCTGCCGGTTCCCCGGCGCCGCCACCCCCGACACGTTCTGGAAG GTCCTGAGCGAGGGCCGGGAGACCCTCACCCACTTCTCCGACGAGGAGCTGCGCGCGGCC GGCGTCGCCGAGCCCCTCCTGGCCGACGACCGTTACGTGAAGGCCGGGCAGGTCCTCGTG GACGCCGACAAGTTCGACGCGGGCCTCTTTGGCATCACCCGGGACGAGGCGGAGCTGATC GACCCCCAGCAGCGCCAGTTCCTGGAGTGCGCGTACGAGGCCCTGGAGCGCGCGGGCTAC GACCCGCAGCGCGGCGAGCAGAGAATCGGCGTCTACGCCGGGGTCGGCCTGAACACCTAT CTGCTGCACAACCTCGGCGAGCGCTACCGCACCGCCTCGTCCGTGGACCGCTACCGCATG ATGATCACGAACGACAAGGACTTCGTCGCCACCCGTACCGCCTACAAGCTGAACCTGTGC GGCCCGAGCGTCAGCACCAACACCGCCTGCTCGACCTCGCTGGTGGCCGTGCACCTGGCC TGCCTCAGCCTGCTCTCCGGCGACTGCACCATGGCGCTCGCGGGCGCCGCCCACATCCAG GCCGACCAGGGCGAGGGCTACCTCCACCACGAGGGGATGATCTTCTCGCCGGACGGCCAC TGCCGGGCCTTCGACGCCAAGGCGCAGGGAACCGTCATCGGCAACGGCGTGGGCGCCGTG GTCCTGAAGCGGCTGAGCGACGCGCTGGCCGACGGCGACACCGTGCACGCCGTCATCAAG GGCACCGCCGTCAACAACGACGGTTCGGACAAGACGGGTTACACCGCGCCGAGCGTCCAG GGACAGGCCGCCGTCGTCGCCGAGGCCCAGGAGATCGCCGACGTCGGCCCGGAGACCGTC TCGTACGTGGAGGCCCACGGCACGGCCACGCCGCTCGGCGACCCGATCGAGGTCGCCGCG CTCAACCAGGCCTTCAACCGCGAGGGTGCCGCACTCGCCCCCGGCTCCTGCGCGCTGGGC TCCGTCAAGACCAACGTCGGCCACCTCGACACGGCCGCGGGCATGGCGGGCCTGATCAAG ACGATCCTGATGCTGCGCCACCGCACCCTGGTCCCGAGCCTCTGCTTCGAAGCCCCCAAC CCGGAGATCGACTTCGCGGCGGGACCCTTCTACGTCGGCACCGAGACCAAGGAGTGGCCC GCGGGCCCGACCCCGCGCCGGGCGGGCGTGAGCTCCTTCGGCATCGGCGGCACCAACGCC CACGTCATCGTGGAGGAGCCGCCCGCCGTCGGCCCCGCCCCCGACGCGGCGGCGGCCGGG CCGAGGCTCCTCGTGCTCTCCGCCAACACCCCCGCCGCGCTGGACACGGCCACCGCCGAC CTCGCCCGCGCGCTCCGCAAGGACCGCGACCTCGACCTGTCCGCCGTGGCCCAGACCCTG GCCCTCGGCCGCCGCGTGCTCCCCTACCGGCGCGCGCTGGTCGCCACCGGCGTACGGGAC GCCGCGCTCGCCCTGGCCCTGGGCGACGCGGGACGGGTCATGACCGCCGGCCCCGCCGAC GAGCGGCCGGTCGTCGAGCTGGTGACCGGCGGCGGCACGCCCGAGCACGCGGCGGCCCTG TACGAGGAGGCGGCGGCCTTCCGCGAGCACTTCGACCGGTGCGCGGCCGAGCTGGGCACG CCCGCCGCCGAACTGCTGCGCGGGCACGGCCCGGACGCGGCCTTCGCCGTCCAGTACGCC ACCGCGCGGGCCCTGGCCGGCTGGGGCAGCACCGCCCCCGTGGTCGCCGCCGACCGCACC GAGCTGCCCGACGCGGCCCTGCGTCTGCTCGACGGGATCGGCGCGCAGCACACCGCCGGC GCCGGACGGCCCGGCGTGGCCCTGCTGCCGGCCGCGTCGGCCCCCGTCGGCACGGCGTTC CTGCTCGGCCTCATCGGGCGGCTGTGGACGGCGGGCGACACCGTCGACTGGACGGTGTTT CACCAGGGCGAGCCGGTACGGAGGGTGCCGCTGCCGACCTACCCCTTCGAGCGCGTACGG CACTGGGCGGAGCCGCACGGCGGCGCGGCACCGGCCGCGGGCCCCGCGGCGCTGACCCTG CGGGACCGCTTCCAGGGAGTGCCGGACGCGGAGAAGAGCGCCCTGCTCACCGGGTTCATC CGGCAGGAGATCGCGGCGGTCATGGGTGCCTCCGACCCGCGGACCGTGGACCTCGACACC AATCTGTTCGACATGGGCCTCGACTCGCTGATCCTGATCGAGGTCATCGCCAAGCTCAGC GACGAACTGGCGCATCCGGTGCGCTCGTCGGCGTTCGTGGAGTACCCGACGGTGCGTTCG TTCGTCGCGGACCTCGGCGAGGAGCTGGGCATCGTCCCGGGGACACCGGAGGCGGCACCC GCCGCCTCCGGGGGCCGGGTGTCCCGCCGGGCGCAGCGGGCCGCCGCCCGCCGCCCCGGG GGTGCGTGA
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR006162 Phosphopantetheine attachment site (PTM)
IPR009081 Acyl carrier protein-like (Domain)
IPR014030 Beta-ketoacyl synthase, N-terminal (Domain)
IPR014031 Beta-ketoacyl synthase, C-terminal (Domain)
IPR016038 Thiolase-like, subgroup (Domain)
IPR016039 Thiolase-like (Domain)
IPR018201 Beta-ketoacyl synthase, active site (Active_site)
IPR020806 Polyketide synthase, phosphopantetheine-binding domain (Domain)
|
| SignalP | No significant hit |
| TMHMM | No significant hit |


 Oxzol_00210
Oxzol_00210


