Salino_00960 : CDS information
 Location
Location
| Organism | Streptomyces albus subsp. albus |
|---|---|
| Strain | DSM 41398 |
| Entry name | Salinomycin |
| Contig | HE586118 |
| Start / Stop / Direction | 154,134 / 158,441 / + [in whole cluster] 154,134 / 158,441 / + [in contig] |
| Location | 154134..158441 [in whole cluster] 154134..158441 [in contig] |
| Type | CDS |
| Length | 4,308 bp (1,435 aa) |
 Annotation
Annotation
| Category | 1.1 PKS |
|---|---|
| Product | polyketide synthase |
| Product (GenBank) | type I modular polyketide synthase |
| Gene | |
| Gene (GenBank) | salAVI |
| EC number | |
| Keyword | |
| Note | |
| Note (GenBank) | |
| Reference |
|
| Related Reference |
|
 PKS/NRPS Module
PKS/NRPS Module
| 10 | 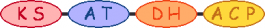 | methylmalonyl-CoA |
 Sequence
Sequence
>polyketide synthase [type I modular polyketide synthase] MANHANNEEKLLEYLKRATADLQQTRKRLQAVQAAEHEPIALVGMACRYPGDADSPEALW DLVAKGQDAVGAFPGNRGWDIESRYSADPDVPGSNYVREGAFLYDVDEFDADFFGINPRE ALAMDPQQRLLLETGWEAVERAGIDPLSLTGSATGTFVGFSAMDYTSGMTEVPEEVEGYI GTGNLASVMSGRLSYSLGLEGPAVTIDTACSSSLVAMHMAAQALRAGECSMALAGGVTVM SNQAGFIEFSRQRALAPDGRCKAFAAAADGFGPSEGAGIVVMERLSDAQRAGRRILAVLR GSAVNQDGASNGLTAPNGPSQVRVIRQALANARVPAAEVDAVEGHGTGTTLGDPIEAQAL LATYGQNRPEGQPLWLGSLKSNIGHTSAASGVASVMKMVMAMRNGLLPQTLHVDAPSPKI DWDSGAVELLTEARDWPRTGRPRRAGVSAFGISGTNAHVILEEAPATEAPAEGAEASDPA ADSASGEAAPALLSRLGTTVPWLLSGRTAKALRGQAERLRRYAATGDLDATATGVALATS RSAFDHRAAVAAPGHEDTLRGLAALAEGDLAAGALQGRDQGGKSVFVFPGQGSQWAGMAV ELLDSSPVFAARLGECAQALDPFVDWSLVDVLRQTEGAPGFDRVDVVQPALWAVMVSLAE VWRAAGVAPAAVIGHSQGEIAAAAVSGALSLSDAAKVSALRARALLALAGKGEMVSVADA AGSVRKRISAWGERLALASVNGPQSTVVSGDPEALDELMAACEAEGVRARRINVDYASHG PQVEQIREEVLGLLDGIQPRAAEVPFLSTVTGEFVTGTELDAEYWYRNLRNTVRFEDAVR QLLTSGHSLFLEASAHPVLTVGVQESIDAAGAAATSQGTLRREEGGPDRLLASLAEAWAH GAAVDWRLVYAGHEASGADLPTYAFQRQRYWLDVAAAAAADVTAAGLGATDHPLLGAAVR LAEGNQTVLTGRLSLRTHPWLGDHVVSGVRVLPGGAFVELAVRAGDEAGCDRLTELALET PMVIPAEGAVQVQVVVGPESDTGRAVTVYGRPESAGDAGEWTRHAHGRLERSDSPEGGTE LTAWPPSGAERVDLDGFYEGLHQQGYGYGPVFQGLADVWLAGDGGGVFAEVTLPAEQQED AGRFGLHPALLDSALHALAVSGPEAEEGTAWLPYAFTGLRLYAAGATRVRVHLKPVGPEE VALVVADVTGRPVATVDSFASRLVSGAELRALADAVAGGTAAAVLPGAPLRRAAAGQDDA EAESFTARLAAMGPEEQERALTDLVRSRVAAVLGHGESETVEAERAFRDLGFDSLTAVEL RNRLTAATGLSLPATLVFSHPTPLALARHLLDQLGTAGAAEGPVAREFAALEAALSAEAP EGEDREAVVGRLEALLWRWRERSGSGSGGVIESGALDSVSDDEIFDLIDKELGAS
>polyketide synthase [type I modular polyketide synthase] ATGGCGAACCACGCGAACAACGAAGAGAAACTGCTCGAATACCTCAAGCGGGCCACGGCC GATCTGCAGCAGACCCGCAAGCGCCTGCAGGCCGTCCAGGCGGCGGAGCACGAGCCCATC GCCCTCGTCGGCATGGCCTGCCGCTACCCCGGCGACGCCGACTCTCCCGAGGCGCTGTGG GACCTGGTGGCCAAGGGCCAGGACGCGGTGGGCGCCTTCCCCGGCAACCGCGGCTGGGAC ATCGAGTCCCGCTACAGCGCGGACCCCGACGTCCCCGGCAGCAACTACGTACGGGAGGGC GCCTTCCTCTACGACGTGGACGAGTTCGACGCCGACTTCTTCGGGATCAACCCGCGTGAG GCGCTCGCCATGGACCCGCAGCAGCGGCTGCTCCTGGAGACCGGCTGGGAGGCCGTGGAG CGCGCGGGCATCGACCCGCTGAGCCTGACCGGCAGCGCCACCGGCACCTTCGTCGGCTTC AGCGCCATGGACTACACCTCGGGCATGACCGAGGTGCCCGAGGAGGTCGAGGGCTACATC GGCACCGGCAACCTCGCCAGCGTCATGTCGGGCCGGCTCTCGTACTCCCTCGGTCTCGAA GGCCCGGCGGTCACCATCGACACCGCCTGCTCCTCCTCGCTCGTCGCCATGCACATGGCC GCGCAGGCGCTGCGGGCGGGGGAGTGCTCCATGGCCCTGGCGGGCGGCGTCACCGTGATG TCCAACCAGGCCGGGTTCATCGAGTTCAGCCGTCAGCGGGCGCTCGCCCCGGACGGCCGC TGCAAGGCCTTCGCCGCCGCGGCCGACGGCTTCGGGCCCTCGGAGGGCGCCGGGATCGTC GTCATGGAGCGCCTCTCCGACGCCCAGCGCGCGGGCCGCCGGATCCTCGCCGTGCTGCGC GGCTCCGCCGTCAACCAGGACGGCGCCTCCAACGGCCTCACCGCCCCCAACGGCCCCTCC CAGGTACGCGTCATCCGGCAGGCCCTCGCCAACGCCCGGGTGCCCGCCGCCGAGGTCGAC GCGGTCGAGGGCCACGGCACCGGCACCACCCTCGGCGACCCCATCGAGGCGCAGGCCCTG CTCGCCACCTACGGCCAGAACCGGCCGGAGGGGCAGCCGCTCTGGCTCGGCTCGCTGAAG TCGAACATCGGCCACACCTCGGCCGCCTCGGGTGTCGCCAGCGTCATGAAGATGGTCATG GCCATGCGCAACGGCCTGCTCCCGCAGACCCTGCACGTGGACGCGCCCTCGCCGAAGATC GACTGGGACTCCGGCGCCGTCGAACTCCTCACCGAGGCCCGCGACTGGCCCCGCACCGGC CGCCCCCGCCGCGCCGGTGTCTCCGCCTTCGGCATCAGCGGCACCAACGCCCACGTCATC CTCGAAGAGGCCCCGGCGACGGAGGCCCCGGCCGAGGGCGCCGAGGCGAGCGACCCCGCC GCGGACTCCGCCAGCGGCGAGGCGGCCCCGGCGCTCCTGTCCCGGCTCGGCACTACGGTG CCCTGGCTGCTCTCCGGACGGACCGCCAAGGCCCTGCGCGGGCAGGCCGAACGGCTCCGC CGGTACGCGGCCACCGGCGACCTCGACGCCACCGCCACCGGTGTCGCCCTCGCCACCTCG CGGTCCGCCTTCGACCACCGCGCCGCCGTCGCCGCCCCCGGCCACGAGGACACCCTCCGC GGCCTGGCGGCCCTCGCCGAGGGCGACCTCGCCGCCGGCGCGCTCCAGGGCCGCGACCAG GGCGGCAAGTCCGTCTTCGTCTTCCCGGGTCAGGGTTCGCAGTGGGCGGGGATGGCGGTC GAACTCCTCGATTCCTCCCCGGTGTTCGCGGCTCGTCTGGGGGAGTGCGCGCAGGCGCTG GACCCCTTCGTGGACTGGTCCCTGGTGGATGTCCTGCGGCAGACCGAGGGCGCCCCCGGC TTCGACCGCGTCGATGTGGTGCAGCCCGCGCTGTGGGCGGTGATGGTGTCGCTTGCCGAG GTGTGGCGTGCGGCGGGTGTCGCCCCGGCCGCGGTGATCGGTCACTCGCAGGGCGAGATC GCGGCTGCGGCGGTGTCCGGTGCGTTGTCCCTGTCGGATGCGGCGAAGGTTTCGGCGTTG CGGGCGCGGGCGTTGCTGGCGCTGGCGGGCAAGGGCGAGATGGTCTCGGTCGCGGATGCG GCCGGTTCCGTACGGAAGCGGATCTCCGCTTGGGGTGAGCGGCTTGCGCTCGCTTCGGTG AACGGTCCGCAGTCGACGGTGGTTTCGGGTGATCCCGAGGCGCTGGACGAGTTGATGGCG GCCTGTGAGGCGGAGGGGGTGCGGGCCCGGCGGATCAATGTGGACTACGCCTCGCACGGC CCCCAGGTCGAGCAGATCCGCGAGGAAGTGCTCGGCCTCCTGGACGGTATTCAGCCACGC GCTGCCGAGGTGCCGTTCCTGTCCACGGTGACTGGTGAGTTCGTGACCGGCACCGAGCTG GACGCCGAGTACTGGTACCGCAACCTGCGCAACACGGTCCGATTCGAGGACGCCGTACGG CAGTTGCTGACCTCGGGGCACAGCCTGTTCCTGGAGGCCAGCGCGCACCCGGTGCTCACC GTGGGCGTGCAGGAGAGCATCGACGCCGCCGGTGCCGCCGCCACCTCGCAGGGCACCCTG CGCCGCGAGGAGGGCGGCCCCGACCGGCTGCTCGCCTCCCTCGCGGAGGCCTGGGCGCAC GGTGCCGCGGTCGACTGGCGCCTGGTCTACGCCGGGCACGAGGCCTCCGGCGCCGACCTG CCCACGTACGCCTTCCAGCGGCAGCGGTACTGGCTGGATGTCGCCGCCGCGGCCGCCGCC GATGTCACCGCGGCGGGGCTCGGCGCCACCGACCACCCCCTGCTCGGCGCCGCGGTCCGG CTCGCCGAGGGCAACCAGACCGTGCTGACCGGGCGGCTCTCGCTGCGCACCCACCCCTGG CTCGGCGACCACGTGGTCTCCGGGGTGCGGGTGCTGCCGGGCGGCGCCTTCGTCGAGCTC GCGGTGCGCGCGGGCGACGAGGCCGGCTGCGACCGGCTCACCGAACTCGCCCTGGAGACC CCGATGGTGATCCCCGCCGAGGGCGCCGTCCAGGTGCAGGTGGTGGTCGGTCCGGAGAGC GACACGGGACGCGCCGTCACCGTCTACGGGCGGCCCGAGAGCGCCGGGGACGCGGGCGAG TGGACCCGGCACGCCCACGGCCGCCTGGAGCGCTCGGACAGCCCCGAGGGCGGCACCGAA CTCACCGCCTGGCCGCCGAGCGGCGCCGAGCGGGTCGACCTCGACGGCTTCTATGAGGGG CTGCACCAGCAGGGGTACGGGTACGGCCCGGTCTTTCAGGGGTTGGCCGACGTCTGGCTC GCGGGCGACGGTGGCGGTGTGTTCGCCGAGGTGACGCTGCCAGCGGAGCAGCAGGAAGAC GCAGGCCGGTTCGGCCTCCACCCGGCCCTGCTCGACTCGGCGCTCCACGCGCTCGCGGTC AGCGGGCCGGAGGCGGAGGAGGGCACGGCCTGGCTGCCGTACGCCTTCACGGGGCTGCGC CTGTACGCGGCCGGTGCCACCCGGGTGCGCGTCCACCTCAAGCCGGTCGGCCCCGAGGAA GTGGCCCTCGTGGTCGCCGATGTGACCGGGCGTCCGGTCGCCACCGTCGACTCCTTCGCC TCCCGGCTGGTCTCCGGCGCCGAGCTGAGGGCCCTCGCGGACGCGGTTGCCGGAGGCACC GCCGCGGCCGTCCTGCCGGGAGCCCCGCTGCGCCGCGCGGCGGCCGGGCAGGACGACGCC GAGGCCGAGTCCTTCACCGCCCGGCTCGCCGCGATGGGCCCCGAGGAGCAGGAGCGCGCG CTGACCGACCTGGTGCGCAGCCGGGTCGCCGCGGTGCTCGGGCACGGCGAGAGCGAGACC GTGGAGGCCGAACGCGCCTTCCGCGACCTCGGCTTCGACTCGCTCACCGCGGTCGAGCTG CGCAACCGGCTCACCGCCGCCACCGGCCTGAGCCTGCCCGCGACGCTGGTCTTCAGCCAC CCCACCCCGCTGGCCCTCGCCCGGCACCTGCTCGACCAGCTCGGCACCGCGGGCGCCGCG GAGGGCCCGGTGGCCCGCGAGTTCGCCGCCCTCGAAGCGGCCCTGTCGGCCGAGGCCCCC GAGGGCGAGGACCGCGAGGCGGTCGTCGGCCGCCTGGAGGCCCTGCTGTGGCGGTGGCGG GAGCGCTCCGGCTCCGGTTCCGGGGGTGTGATCGAGTCCGGCGCCCTCGACTCGGTGTCC GACGACGAGATCTTCGACCTGATCGACAAGGAACTGGGAGCTTCCTGA
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR001227 Acyl transferase domain (Domain)
IPR006162 Phosphopantetheine attachment site (PTM)
IPR009081 Acyl carrier protein-like (Domain)
IPR014030 Beta-ketoacyl synthase, N-terminal (Domain)
IPR014031 Beta-ketoacyl synthase, C-terminal (Domain)
IPR014043 Acyl transferase (Domain)
IPR015083 Polyketide synthase, docking (Domain)
IPR016035 Acyl transferase/acyl hydrolase/lysophospholipase (Domain)
IPR016036 Malonyl-CoA ACP transacylase, ACP-binding (Domain)
IPR016038 Thiolase-like, subgroup (Domain)
IPR016039 Thiolase-like (Domain)
IPR018201 Beta-ketoacyl synthase, active site (Active_site)
IPR020801 Polyketide synthase, acyl transferase domain (Domain)
IPR020806 Polyketide synthase, phosphopantetheine-binding domain (Domain)
IPR020807 Polyketide synthase, dehydratase domain (Domain)
IPR020841 Polyketide synthase, beta-ketoacyl synthase domain (Domain)
|
| SignalP | No significant hit |
| TMHMM | No significant hit |


 Salino_00950
Salino_00950


