Azino_00250 : CDS information
 Location
Location
| Organism | Streptomyces sahachiroi |
|---|---|
| Strain | NRRL 2485 (=NBRC 13928) |
| Entry name | Azinomycin B |
| Contig | EU240558 |
| Start / Stop / Direction | 33,636 / 38,261 / + [in whole cluster] 33,636 / 38,261 / + [in contig] |
| Location | 33636..38261 [in whole cluster] 33636..38261 [in contig] |
| Type | CDS |
| Length | 4,626 bp (1,541 aa) |
 Annotation
Annotation
| Category | 1.2 NRPS |
|---|---|
| Product | non-ribosomal peptide synthetase |
| Product (GenBank) | Azi25 |
| Gene | aziA5 |
| Gene (GenBank) | azi25 |
| EC number | |
| Keyword | |
| Note | |
| Note (GenBank) |
|
| Reference |
|
 PKS/NRPS Module
PKS/NRPS Module
| B | 5 | 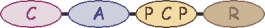 | threonine |
|---|
 Sequence
Sequence
>non-ribosomal peptide synthetase [Azi25] MDLPTEQPRSLPDTPVAAAVPPDLAALRSRLAELSPEKRRLVERLLARQGAGLGITRRAG GAPEAPCSFEQERLWFMYELLTRREIFHVPVALRLEGELDPDALERALRQLGRRHEALRT VFRQRDGRPYQVVREQLDLPLERVDCRTAADPALAARQQASALVTEDFDLEEGPLVRCTL YRTGEHEHLLAVVQHHIVSDNWSLGILLDDLGKLYARELGAPGELPPLDLHYPDFAAWQR ATVDSTTMRRTLDHWRDRLDGAPDSLDLPTDRPRPAVRGSQGKFHHVRFDADLVAGLREA ARQHDTTLLGAFLAGYIALLSRLVRSDSLVVGVPVAGRPRAEMQQMIGYFLNWLPIHVQV GDRPDLHTLIRRTGTALSEAMTHQDVPFDMLVRELRPSRRPGETPIFQTSFSLRDGAPTP PAMPGIDVTFAELDGGATHYDLMAELWCEGDEVVGYVPYDDELLDAQTVARWAGWLKTLL RAGLAAPDTPVADLEMLAPGESVVIPARAPVTAEGGVARPARTLHGVFAAQAARRPDAPA VSDERDRLTYAELSARADRIAAALQERGEGPGSIVGLVLDRTVDLPAAVLGVLRAGAAYL PVDPENPAGRTADQFTECRVRTVLTTPATASAPALEELDDGPRILVVDWQDPGWREQTPG PAAVDVPADAPAYVIYTSGSTGRPKGVLVTHRNVLRLFTACDEHLRVGPDDVWTLFHSYA FDFSVWEIWGALLHGGRLVVVPQWATRAPDVFAELVRDERVTVLSQTPSAFGQVSETLLK HPDPAALRYVVFGGEALDPTALRPWTRVYGDRRPELINMYGITETTVHVTARPLREKDLA GTASPIGPPLADLSLYLLDDSLRPVGTGVPGEIFVGGDGVSLGYVANPGLTAERMLPDPF AGRPGARMYRSGDLAVRRGDGELVCLGRADDQVKIRGHRIEPGEVRAALDALAVVARAAV VVERDRVGAAVLAAYVVPAEGNAGVSGTGIRRTLLRSLPEWMVPASVTVLDELPLTRNGK LDRRALTDRKEHAAPAGPRGEAPRSDTARQLAEIWQELLGVPAVGGEDSFFELGGHSLMV MHLVARIRTAFGVEMPVETLFRRPQLQPLADEVDAARTAAQRAAAPEPGQDAAAELADVR ADLAARAAGIPRPGARADADRDTVLLTGATGFVGRFVLAELLAAGARVICLLRGGTARRE ELVAGMADLGLWHEEHAARLELVDGDIAEPGLGLAGPDRDRLADRAGRIIHAAAWVNHVY PYERLAAANTHCMAGLLELAARGRRSALTVVSTSSVADSAAYPPGSTVPPGPLKALPSAA NGYVRSKAVAEQYLHLAAELDVPAAVIRIPSVFGDQRRYQINPADAVWSWCRAMIETSGF PESFAQPGNELFQALPADAVARAVLLADRDHTEPGTRYLDAVPAAVGTTEDLLAALRAAG HALSPCPDHAWYTAVGELDPGRVWVAGIAGQAAARLAADPSATAPRTLRRFTAPVEPGEL DELLRTRALYSSAQLAGYIRTLDASFPPAGNDREKARPARG
>non-ribosomal peptide synthetase [Azi25] ATGGATCTGCCCACCGAGCAGCCACGGTCCCTGCCGGACACGCCGGTGGCCGCCGCGGTG CCGCCCGATCTCGCCGCCCTGCGCTCGCGGCTGGCCGAACTGAGCCCGGAAAAACGCCGG TTGGTGGAGCGACTGCTGGCCCGTCAGGGCGCCGGCCTCGGCATCACCCGTCGCGCCGGC GGTGCGCCCGAGGCCCCCTGCTCCTTCGAGCAAGAGCGGCTGTGGTTCATGTACGAGCTG CTGACCCGGCGCGAGATCTTTCACGTCCCGGTCGCCCTGCGCCTCGAAGGCGAACTGGAC CCCGACGCGCTGGAGCGGGCACTGCGGCAGCTCGGCCGACGGCACGAGGCACTGCGCACC GTGTTCCGGCAGCGCGACGGCCGCCCCTACCAGGTGGTACGGGAACAGCTCGATCTCCCG CTGGAGCGGGTCGACTGCCGCACCGCCGCCGATCCCGCCCTCGCCGCCCGGCAGCAGGCC TCCGCACTGGTCACCGAGGACTTCGACCTCGAAGAAGGACCCCTGGTGCGCTGCACGCTG TACCGGACGGGGGAACACGAACACCTGCTGGCCGTGGTGCAGCACCACATCGTCTCCGAC AACTGGTCGCTGGGCATCCTGCTCGACGACCTCGGCAAGCTGTACGCCCGCGAGCTGGGG GCACCGGGGGAGCTGCCCCCGCTCGACCTGCACTACCCCGACTTCGCCGCATGGCAGCGG GCCACGGTCGACAGCACGACGATGCGGCGCACCCTCGATCACTGGCGCGACCGCCTGGAC GGCGCCCCGGACAGTCTCGACCTGCCCACCGACCGTCCCCGGCCCGCGGTCCGCGGCAGC CAGGGGAAGTTCCACCACGTACGGTTCGACGCCGACCTGGTCGCCGGGCTGCGCGAAGCG GCCCGGCAGCACGACACCACCCTCCTGGGCGCGTTCCTCGCAGGGTACATAGCCCTGCTC TCCCGCCTCGTGCGGTCGGACTCGCTGGTGGTCGGTGTACCGGTGGCCGGGCGGCCGCGC GCGGAGATGCAGCAGATGATCGGGTACTTCCTGAACTGGCTGCCGATCCACGTCCAGGTC GGTGACCGGCCGGACCTGCACACCCTGATACGCCGCACCGGAACCGCCCTGAGCGAGGCG ATGACCCATCAGGACGTGCCGTTCGACATGCTGGTGCGCGAGCTGCGGCCCTCGCGGCGC CCCGGGGAGACACCGATCTTCCAGACCTCCTTCTCGCTGCGTGACGGCGCCCCCACCCCG CCGGCCATGCCGGGCATCGACGTCACCTTCGCCGAACTCGACGGCGGAGCCACCCATTAC GACCTGATGGCCGAACTGTGGTGCGAGGGCGACGAGGTCGTGGGCTACGTGCCCTACGAC GACGAACTGCTGGACGCGCAGACGGTGGCCCGCTGGGCCGGCTGGCTCAAGACGCTGCTG CGCGCCGGGCTGGCCGCACCGGACACCCCGGTGGCGGACCTGGAGATGCTCGCTCCCGGC GAGTCCGTGGTGATCCCGGCCCGCGCGCCGGTGACGGCCGAGGGGGGCGTCGCCCGGCCG GCCCGGACCCTGCACGGGGTGTTCGCCGCGCAGGCGGCTCGGCGGCCCGACGCCCCCGCC GTCTCCGACGAGCGGGACCGGCTCACCTACGCGGAACTGTCCGCACGAGCCGACCGGATC GCCGCCGCCCTGCAAGAGCGGGGTGAGGGACCCGGCAGCATCGTCGGCCTCGTCCTCGAC CGCACCGTGGACCTGCCCGCCGCCGTTCTCGGCGTGCTTCGCGCGGGTGCCGCCTACCTG CCCGTCGACCCGGAGAATCCCGCCGGACGCACCGCTGACCAGTTCACCGAGTGCCGGGTC CGGACCGTGCTGACCACCCCGGCCACCGCGTCGGCACCCGCGTTGGAGGAACTGGACGAC GGACCGCGCATCCTCGTGGTGGACTGGCAGGACCCCGGATGGCGGGAGCAGACCCCCGGC CCGGCTGCCGTGGACGTACCCGCGGACGCGCCCGCCTACGTCATCTACACCTCGGGATCC ACCGGCCGCCCCAAGGGCGTGCTCGTCACGCACCGCAACGTGCTCCGGCTGTTCACCGCC TGCGACGAGCATCTGAGGGTGGGGCCGGACGACGTATGGACCCTGTTCCACTCCTACGCC TTCGACTTCTCCGTCTGGGAGATCTGGGGTGCGCTGCTGCACGGCGGCAGGCTGGTGGTC GTCCCGCAGTGGGCGACCCGCGCCCCCGACGTCTTCGCCGAACTCGTACGGGACGAGCGG GTCACCGTGCTGAGCCAGACCCCGTCCGCGTTCGGCCAGGTGAGCGAGACCCTGCTGAAG CACCCCGACCCGGCGGCGCTGCGGTACGTGGTGTTCGGCGGCGAGGCACTCGATCCCACC GCGCTGCGTCCCTGGACGCGTGTGTACGGCGACCGGCGGCCGGAACTGATCAACATGTAC GGGATCACCGAGACCACCGTGCACGTCACCGCGCGGCCGCTGCGCGAGAAGGACCTCGCC GGGACGGCGTCCCCGATCGGACCTCCGCTCGCCGACCTGTCCCTGTACCTGCTGGACGAT TCCCTGCGTCCGGTCGGCACCGGTGTACCCGGGGAGATCTTCGTCGGAGGGGACGGGGTG AGCCTGGGCTACGTCGCCAACCCGGGGCTGACCGCGGAGCGCATGCTCCCCGACCCGTTC GCCGGACGTCCCGGCGCCCGTATGTACCGCAGCGGCGACCTGGCGGTCCGCCGCGGCGAC GGAGAGCTGGTCTGTCTGGGCCGCGCCGACGACCAGGTGAAGATCCGCGGGCACCGGATC GAGCCGGGTGAGGTCCGGGCGGCGCTGGACGCCCTGGCCGTCGTCGCCCGTGCCGCCGTG GTGGTGGAACGCGACCGGGTGGGCGCCGCCGTCCTGGCGGCCTACGTCGTACCGGCCGAG GGGAACGCCGGCGTCAGCGGCACCGGGATTCGCCGGACCCTGCTGCGCTCCCTGCCGGAA TGGATGGTGCCCGCCTCGGTCACCGTCCTCGACGAGCTGCCCCTGACCCGGAACGGCAAG CTGGACCGCCGGGCCCTGACCGACCGCAAGGAGCACGCGGCGCCGGCCGGACCGCGCGGC GAGGCGCCCCGCAGCGACACCGCGCGGCAACTTGCCGAGATCTGGCAGGAGCTGCTCGGC GTCCCCGCGGTGGGCGGCGAGGACAGCTTCTTCGAACTCGGCGGCCACTCCCTGATGGTG ATGCACCTCGTCGCCCGGATACGCACGGCCTTCGGCGTCGAGATGCCGGTGGAGACCCTC TTCCGCCGCCCTCAACTGCAGCCGCTGGCCGACGAGGTGGACGCGGCACGTACCGCGGCG CAGAGGGCCGCGGCGCCGGAGCCGGGACAGGACGCGGCGGCCGAACTCGCCGACGTGCGC GCCGACCTGGCCGCCCGGGCCGCCGGGATCCCCCGCCCGGGGGCCCGGGCGGACGCGGAC CGCGACACAGTCCTGCTCACCGGGGCCACGGGCTTCGTCGGCCGGTTCGTGCTCGCCGAA CTCCTTGCCGCGGGGGCCCGGGTGATCTGCCTGCTCCGCGGCGGGACCGCCCGCCGGGAG GAGCTGGTGGCGGGGATGGCGGACCTCGGGCTGTGGCACGAGGAGCACGCCGCACGGCTG GAACTGGTCGACGGCGACATCGCCGAACCCGGGCTCGGCCTCGCCGGACCGGACCGCGAC CGCCTGGCCGACCGGGCGGGCCGCATCATCCACGCCGCGGCCTGGGTGAACCACGTCTAC CCGTACGAGCGGCTGGCCGCGGCGAACACGCACTGCATGGCAGGTCTGCTGGAACTCGCG GCCCGCGGCCGCCGCTCGGCTCTCACTGTGGTGTCCACCAGCTCGGTGGCCGACTCCGCC GCGTACCCGCCCGGCTCGACCGTGCCGCCGGGCCCGCTGAAGGCACTGCCGTCCGCCGCG AACGGATACGTGCGTTCCAAGGCGGTGGCCGAGCAGTACCTCCACCTGGCAGCCGAACTG GACGTACCCGCCGCGGTGATCAGGATTCCCAGCGTCTTCGGCGATCAGCGGCGGTACCAG ATCAACCCCGCAGACGCCGTGTGGAGTTGGTGCCGGGCGATGATCGAGACCAGCGGCTTC CCCGAGAGCTTCGCGCAGCCGGGCAACGAACTGTTCCAGGCGCTGCCCGCCGACGCCGTG GCCCGGGCGGTGCTGCTGGCCGACCGCGATCACACCGAACCGGGCACCCGCTACCTGGAC GCCGTCCCGGCCGCCGTCGGCACGACAGAGGACCTGCTGGCCGCTCTGCGCGCCGCCGGT CATGCACTGAGCCCCTGCCCGGACCACGCGTGGTACACCGCCGTGGGCGAGCTGGACCCC GGCCGGGTCTGGGTGGCGGGCATCGCAGGGCAGGCCGCCGCCCGGCTGGCCGCGGATCCG TCCGCCACGGCCCCCCGCACCCTGCGCCGCTTCACCGCCCCCGTGGAGCCCGGTGAGCTG GACGAGCTGTTGCGCACCCGGGCCCTGTACTCCTCGGCCCAGCTGGCCGGGTACATCCGG ACGCTCGACGCGTCCTTCCCGCCCGCCGGCAACGACCGGGAGAAAGCCCGGCCCGCGCGC GGCTGA
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR000873 AMP-dependent synthetase/ligase (Domain)
IPR001242 Condensation domain (Domain)
IPR006162 Phosphopantetheine attachment site (PTM)
IPR009081 Acyl carrier protein-like (Domain)
IPR010071 Amino acid adenylation domain (Domain)
IPR013120 Male sterility, NAD-binding (Domain)
IPR016040 NAD(P)-binding domain (Domain)
IPR020845 AMP-binding, conserved site (Conserved_site)
IPR025110 Domain of unknown function DUF4009 (Domain)
|
| SignalP | |
| TMHMM | No significant hit |


 Azino_00240
Azino_00240


