Calicheamicin : Cluster information
 Compound
Compound
| Entry name | Calicheamicin |
|---|---|
| Moiety description | A: enediyne core B: orsellinic acid moiety |
| PKS Type | A: TypeI iterative B: TypeI iterative |
| Starter Unit | A: acetyl-CoA B: acetyl-CoA |
| Chain Length | A: 8 B: 4 |
| Sugar Unit | 3-methoxy-L-rhamnose aminodideoxypentose 4-hydroxyamino-6-deoxy-alpha-D-glucose 4-thio-2,4,6-trideoxy-D-glucose |
| Classification | Enediyne |
| Activity | Antibiotic Antitumor |
| Composition | γ1I: C55H74IN3O21S4 |
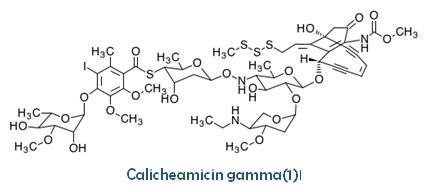
 Original source
Original source
| Organism | Micromonospora echinospora |
|---|---|
| Strain | NRRL 15839 |
| Contig | AF497482 |
 PKS/NRPS Module
PKS/NRPS Module
| A | Calm_00360 calE8 | 1 |  | acetyl-CoA malonyl-CoA |
|---|---|---|---|---|
| B | Calm_00640 calO5 | 1 |  | acetyl-CoA malonyl-CoA |
 Reference
Reference
- The calicheamicin gene cluster and its iterative type I enediyne PKS.
- Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS[PMID: 12183629]Science. 297 (2002) 1173-6
- On the origin of deoxypentoses: evidence to support a glucose progenitor in the biosynthesis of calicheamicin.
- Bililign T, Shepard EM, Ahlert J, Thorson JS[PMID: 12404643]Chembiochem. 3 (2002) 1143-6
- A genomics-guided approach for discovering and expressing cryptic metabolic pathways.
- Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM[PMID: 12536216]Nat Biotechnol. 21 (2003) 187-90
- Resistance to enediyne antitumor antibiotics by CalC self-sacrifice.
- Biggins JB, Onwueme KC, Thorson JS[PMID: 12970566]Science. 301 (2003) 1537-41
- Deciphering indolocarbazole and enediyne aminodideoxypentose biosynthesis through comparative genomics: insights from the AT2433 biosynthetic locus.
- Gao Q, Zhang C, Blanchard S, Thorson JS[PMID: 16873021]Chem Biol. 13 (2006) 733-43
- Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions.
- Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS[PMID: 16946071]Science. 313 (2006) 1291-4
- Structural insight into the self-sacrifice mechanism of enediyne resistance.
- Singh S, Hager MH, Zhang C, Griffith BR, Lee MS, Hallenga K, Markley JL, Thorson JS[PMID: 17168523]ACS Chem Biol. 1 (2006) 451-60
- Evidence for a novel phosphopantetheinyl transferase domain in the polyketide synthase for enediyne biosynthesis.
- Murugan E, Liang ZX[PMID: 18319060]FEBS Lett. 582 (2008) 1097-103
- Characterization of a carbonyl-conjugated polyene precursor in 10-membered enediyne biosynthesis.
- Kong R, Goh LP, Liew CW, Ho QS, Murugan E, Li B, Tang K, Liang ZX[PMID: 18529057]J Am Chem Soc. 130 (2008) 8142-3
- Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
- McCoy JG, Johnson HD, Singh S, Bingman CA, Lei IK, Thorson JS, Phillips GN Jr[PMID: 18561189]Proteins. 74 (2009) 50-60
- Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
- Zhang C, Bitto E, Goff RD, Singh S, Bingman CA, Griffith BR, Albermann C, Phillips GN Jr, Thorson JS[PMID: 18721755]Chem Biol. 15 (2008) 842-53
- Cloning and characterization of CalS7 from Micromonospora echinospora sp. calichensis as a glucose-1-phosphate nucleotidyltransferase.
- Simkhada D, Oh TJ, Kim EM, Yoo JC, Sohng JK[PMID: 18807197]Biotechnol Lett. 31 (2009) 147-53
- Characterization of CalE10, the N-oxidase involved in calicheamicin hydroxyaminosugar formation.
- Johnson HD, Thorson JS[PMID: 19055330]J Am Chem Soc. 130 (2008) 17662-3
- Natural-product sugar biosynthesis and enzymatic glycodiversification.
- Thibodeaux CJ, Melancon CE 3rd, Liu HW[PMID: 19058170]Angew Chem Int Ed Engl. 47 (2008) 9814-59
- Characterization of CalS9 in the biosynthesis of UDP-xylose and the production of xylosyl-attached hybrid compound.
- Simkhada D, Oh TJ, Pageni BB, Lee HC, Liou K, Sohng JK[PMID: 19290519]Appl Microbiol Biotechnol. 83 (2009) 885-95
- Structure and catalytic mechanism of the thioesterase CalE7 in enediyne biosynthesis.
- Kotaka M, Kong R, Qureshi I, Ho QS, Sun H, Liew CW, Goh LP, Cheung P, Mu Y, Lescar J, Liang ZX[PMID: 19357082]J Biol Chem. 284 (2009) 15739-49
- Production of octaketide polyenes by the calicheamicin polyketide synthase CalE8: implications for the biosynthesis of enediyne core structures.
- Belecki K, Crawford JM, Townsend CA[PMID: 19689130]J Am Chem Soc. 131 (2009) 12564-6
- Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
- Chang A, Singh S, Bingman CA, Thorson JS, Phillips GN Jr[PMID: 21358050]Acta Crystallogr D Biol Crystallogr. 67 (2011) 197-203
- Solution structures of the acyl carrier protein domain from the highly reducing type I iterative polyketide synthase CalE8.
- Lim J, Kong R, Murugan E, Ho CL, Liang ZX, Yang D[PMID: 21674045]PLoS One. 6 (2011) e20549
- Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
- Chang A, Singh S, Helmich KE, Goff RD, Bingman CA, Thorson JS, Phillips GN Jr[PMID: 21987796]Proc Natl Acad Sci U S A. 108 (2011) 17649-54
- Rigidifying acyl carrier protein domain in iterative type I PKS CalE8 does not affect its function.
- Lim J, Sun H, Fan JS, Hameed IF, Lescar J, Liang ZX, Yang D[PMID: 23009853]Biophys J. 103 (2012) 1037-44
- Environmental control of the calicheamicin polyketide synthase leads to detection of a programmed octaketide and a proposal for enediyne biosynthesis.
- Belecki K, Townsend CA[PMID: 23042574]Angew Chem Int Ed Engl. 51 (2012) 11316-9
- Structural and Functional Characterization of CalS11, a TDP-Rhamnose 3'-O-Methyltransferase Involved in Calicheamicin Biosynthesis.
- Singh S, Chang A, Helmich KE, Bingman CA, Wrobel RL, Beebe ET, Makino S, Aceti DJ, Dyer K, Hura GL, Sunkara M, Morris AJ, Phillips GN Jr, Thorson JS[PMID: 23662776]ACS Chem Biol. 8 (2013) 1632-9
- Biochemical determination of enzyme-bound metabolites: preferential accumulation of a programmed octaketide on the enediyne polyketide synthase CalE8.
- Belecki K, Townsend CA[PMID: 24041368]J Am Chem Soc. 135 (2013) 14339-48
- A general NMR-based strategy for the in situ characterization of sugar-nucleotide-dependent biosynthetic pathways.
- Singh S, Peltier-Pain P, Tonelli M, Thorson JS[PMID: 24911465]Org Lett. 16 (2014) 3220-3
- Characterization of the calicheamicin orsellinate C2-O-methyltransferase CalO6.
- Singh S, Nandurkar NS, Thorson JS[PMID: 24978950]Chembiochem. 15 (2014) 1418-21
- Enediyne polyketide synthases stereoselectively reduce the beta-ketoacyl intermediates to beta-D-hydroxyacyl intermediates in enediyne core biosynthesis.
- Ge HM, Huang T, Rudolf JD, Lohman JR, Huang SX, Guo X, Shen B[PMID: 25019332]Org Lett. 16 (2014) 3958-61
- Structure-guided functional characterization of enediyne self-sacrifice resistance proteins, CalU16 and CalU19.
- Elshahawi SI, Ramelot TA, Seetharaman J, Chen J, Singh S, Yang Y, Pederson K, Kharel MK, Xiao R, Lew S, Yennamalli RM, Miller MD, Wang F, Tong L, Montelione GT, Kennedy MA, Bingman CA, Zhu H, Phillips GN Jr, Thorson JS[PMID: 25079510]ACS Chem Biol. 9 (2014) 2347-58
- Identification and characterization of a methionine gamma-lyase in the calicheamicin biosynthetic cluster of Micromonospora echinospora.
- Song H, Xu R, Guo Z[PMID: 25404066]Chembiochem. 16 (2015) 100-9
- Structural Basis for the Stereochemical Control of Amine Installation in Nucleotide Sugar Aminotransferases.
- Wang F, Singh S, Xu W, Helmich KE, Miller MD, Cao H, Bingman CA, Thorson JS, Phillips GN Jr[PMID: 26023720]ACS Chem Biol. 10 (2015) 2048-56
- Structural Characterization of CalS8, a TDP-alpha-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.
- Singh S, Michalska K, Bigelow L, Endres M, Kharel MK, Babnigg G, Yennamalli RM, Bingman CA, Joachimiak A, Thorson JS, Phillips GN Jr[PMID: 26240141]J Biol Chem. 290 (2015) 26249-58
- Characterization of Early Enzymes Involved in TDP-Aminodideoxypentose Biosynthesis en Route to Indolocarbazole AT2433.
- Peltier-Pain P, Singh S, Thorson JS[PMID: 26289554]Chembiochem. 16 (2015) 2141-6
- Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
- Han L, Singh S, Thorson JS, Phillips GN Jr[PMID: 26958582]Struct Dyn. 3 (2016) 012004
 Data download
Data download
 History
History
- 2016-12-27[Update]
- 2016-01-27[Update]
- 2015-02-12[Update]
- 2014-06-25[Update]
- 2013-12-27[Release]






