Calm_00640 : CDS information
 Location
Location
| Organism | Micromonospora echinospora |
|---|---|
| Strain | NRRL 15839 |
| Entry name | Calicheamicin |
| Contig | AF497482 |
| Start / Stop / Direction | 76,231 / 80,046 / + [in whole cluster] 76,231 / 80,046 / + [in contig] |
| Location | 76231..80046 [in whole cluster] 76231..80046 [in contig] |
| Type | CDS |
| Length | 3,816 bp (1,271 aa) |
 Annotation
Annotation
| Category | 1.1 PKS |
|---|---|
| Product | polyketide synthase putative orsellinic acid synthase |
| Product (GenBank) | CalO5 |
| Gene | |
| Gene (GenBank) | calO5 |
| EC number | |
| Keyword |
|
| Note | |
| Note (GenBank) | |
| Reference |
|
| Related Reference |
|
|
 PKS/NRPS Module
PKS/NRPS Module
| B | 1 | 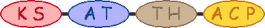 | acetyl-CoA malonyl-CoA |
|---|
 Sequence
Sequence
>polyketide synthase [CalO5] MTRDDPADNPYQVAVIGIGCRLPSDVDTPDALWELLLKGGQTAGEIPAQRWRAYRERGPE YEAVLRDTVTAGSYLRDVAGFDPEFFGLSPREAAEMDPQQRILLEVGWEALEHAGLPPTR LAGTDTGVFVGVSTTDYGDRLLEDLPTVEAYTGIGAATCALANRISYALDLRGPSVAVDT ACSASLVAVHLACQSLLLRESTVALAGGVNLVLTPGQNASLQAAGTLSPDGVSKSFDRDA DGYGRGEGCGVLVLKRLDEAERDGDRVLAVIRGSAVNQDGHTDGIMAPSGPAQQHVVRRA CDRAGVAPASVDYVEAHGTGTQLGDPVEATALAAVYGAGRPDDRPCLIGSIKSNVGHLEG AAGVAGLMKAILALHRGQIPGTPLRGQAIPAVGDGTGLRLVTGTLPWPERDGPRRAAVSG FGYGGTIAHVVLEEAPPVPADGDAPAADEAARPLFPLSARSDAALRQQAARLADRLDGDD RPDLVDVGHTLARRRAHLTHRAVVTGQDRDEVVAALRRLAAGQPDPDTVSAVTPAGRSAR PVWVFSGHGSHWAGMGRDLLAHEPAFAAAIDEIEPVVAEEAGFSLRQALADPELAGVDRI QTLTFAMHVGLAAVWRAYGVRPAAVIGHSVGEVAAAVTAGILTPTDGARLICRRSALLRR AAGRGAMAMVTLPFADVAERLAGRADLVAAIASAPASTVVSGDIAAVDAVIEQWPADRIA VRRVQSDVAFHSPHMDALADELRAAAADLPTSAGTVPMYSTVLDDPRAAAPCDGAYWAAN LRRPVRLVDAVGAALADGHRTFVEISAHPVVAHSVRETLDHAGIEDGYVGTTLRRHVPGH RTALAAVAEAHCHGVEVDWAALRPDGRLTDLPRYAWRHQELWREPTPPVAAGGHDIGSHT LLGTATTVAGSDLRLWHTVLSDASRPYPGSHTVQGAELVPAAVLAATFLAAGARDGAPVA LHDLSMRVPLATAGRQEIQVVDDGGKLLLASRPAGADTAPWLTHATARVTPDDAAPSARA ADPRGPGGVVADPGLVRDRLARVGVPETGFPWRVDRLTTGNGGLRARVRFPEPAEDWAAV IDAAVSIAPAAFPGEPRLRLVDAIERIWTTGVVPAVALVDVVRDGGREDTVDVTVSGADG TLAARLTGLHYPVVPDAPAADTDGPAPTRQSLADLDPDELRERLIEEVRAAVAAEMKLAA EALDPRLPLVQQGMDSVMTVIVRRRLEKTYRQVLPASLFWQQPTVTAIAVHLAELIAAPS PDGAVADAVAR
>polyketide synthase [CalO5] ATGACCCGGGACGATCCCGCCGACAACCCGTACCAGGTGGCCGTCATCGGCATCGGTTGC CGGCTGCCCAGCGACGTCGACACCCCGGACGCCCTCTGGGAGCTGCTACTCAAGGGCGGC CAGACCGCCGGCGAGATCCCGGCGCAGCGCTGGCGCGCCTACCGGGAGCGCGGCCCCGAG TACGAGGCGGTCCTGCGCGACACCGTCACCGCCGGCAGCTACCTGCGTGACGTCGCGGGC TTCGACCCCGAGTTCTTCGGCCTGTCGCCCCGGGAGGCGGCCGAGATGGACCCGCAGCAG CGGATCCTGCTCGAGGTCGGCTGGGAGGCCCTGGAGCACGCCGGCCTGCCACCCACCCGG CTGGCCGGCACCGACACGGGCGTCTTCGTCGGGGTCAGCACCACCGACTACGGCGACCGG CTGCTGGAGGACCTGCCGACCGTCGAGGCGTACACCGGGATCGGCGCGGCCACCTGCGCC CTGGCCAACCGCATCTCCTACGCGCTGGACCTGCGCGGCCCGAGCGTCGCCGTCGACACG GCCTGCTCGGCGTCGCTGGTCGCGGTGCACCTGGCCTGCCAGAGCCTGCTGCTGCGGGAG AGCACGGTGGCCCTGGCCGGCGGCGTGAACCTGGTGCTCACCCCGGGGCAGAACGCCTCC CTCCAGGCCGCCGGCACGCTCTCCCCCGACGGCGTGAGCAAGTCGTTCGACCGCGACGCC GACGGGTACGGCCGCGGCGAGGGATGCGGCGTGCTGGTGCTCAAGCGGCTCGACGAGGCC GAACGGGACGGCGACCGGGTCCTGGCGGTGATCCGGGGCAGCGCGGTCAACCAGGACGGC CACACCGACGGCATCATGGCCCCGTCCGGCCCCGCGCAGCAGCACGTCGTCCGCCGCGCC TGCGACCGGGCGGGCGTCGCCCCGGCCAGCGTGGACTACGTGGAGGCGCACGGCACCGGC ACCCAGCTCGGCGATCCCGTCGAGGCGACCGCCCTGGCCGCGGTCTACGGCGCCGGCCGC CCCGACGACCGGCCCTGCCTCATCGGCTCGATCAAGTCCAACGTCGGGCACCTGGAGGGC GCCGCCGGGGTCGCGGGGCTGATGAAGGCCATCCTCGCGCTGCACCGCGGCCAGATCCCG GGCACCCCGCTGCGCGGCCAGGCGATCCCCGCCGTCGGCGACGGCACCGGGCTGCGCCTG GTGACCGGCACGCTGCCCTGGCCCGAGCGGGACGGTCCGCGCCGCGCCGCGGTCTCCGGG TTCGGCTACGGCGGCACCATCGCCCACGTCGTCCTGGAGGAGGCGCCGCCCGTTCCCGCC GACGGCGACGCGCCGGCTGCCGACGAGGCCGCCCGGCCGCTGTTCCCGCTGTCGGCCCGA TCGGACGCCGCCCTGCGCCAGCAGGCCGCCCGACTCGCCGACCGGCTCGACGGGGACGAC CGGCCGGACCTGGTCGACGTCGGCCACACGCTCGCGCGCCGCCGGGCCCACCTGACCCAC CGCGCGGTGGTCACCGGGCAGGACCGCGACGAGGTCGTCGCCGCCCTGCGCCGGCTCGCC GCCGGCCAACCCGACCCCGACACGGTCTCGGCAGTGACGCCGGCCGGCCGGTCCGCCCGG CCCGTCTGGGTCTTCTCCGGGCACGGCTCGCACTGGGCCGGCATGGGCCGCGACCTGCTG GCCCACGAACCGGCCTTCGCCGCCGCGATCGACGAGATCGAACCGGTCGTCGCCGAGGAG GCCGGCTTCTCGCTGCGGCAGGCCCTCGCCGACCCCGAGCTGGCCGGGGTGGACCGCATC CAGACCCTGACCTTCGCCATGCACGTCGGCCTGGCCGCGGTCTGGCGGGCGTACGGTGTC CGCCCGGCCGCCGTCATCGGGCACTCGGTCGGCGAGGTCGCCGCCGCCGTGACCGCGGGC ATCCTCACCCCGACCGACGGGGCCCGGCTGATCTGCCGCCGGTCCGCCCTGCTGCGGCGG GCCGCCGGCCGGGGCGCGATGGCCATGGTCACCCTGCCGTTCGCCGACGTCGCCGAGCGG CTCGCCGGCCGCGCCGACCTCGTCGCGGCGATCGCCTCCGCCCCCGCGTCCACCGTGGTC TCCGGCGACATCGCCGCCGTGGACGCCGTGATCGAGCAGTGGCCGGCGGACCGGATCGCG GTGCGCCGGGTGCAGTCCGACGTGGCGTTCCACAGCCCGCACATGGACGCCCTCGCCGAC GAGCTGCGGGCCGCCGCGGCCGACCTGCCCACCTCGGCGGGCACCGTGCCGATGTACTCG ACGGTGCTCGACGACCCGCGCGCCGCGGCCCCCTGCGACGGGGCGTACTGGGCGGCGAAC CTGCGCCGACCGGTCCGCCTGGTCGACGCGGTCGGCGCCGCCCTGGCCGACGGGCACCGC ACCTTCGTCGAGATCTCCGCCCACCCGGTGGTCGCCCACTCTGTGCGGGAGACCCTCGAC CACGCCGGCATCGAGGACGGGTACGTCGGGACCACGCTGCGCCGTCACGTTCCCGGGCAC CGCACGGCCCTCGCCGCGGTGGCGGAGGCGCACTGCCACGGCGTCGAGGTCGACTGGGCC GCGCTGCGGCCCGACGGCAGGCTCACCGACCTGCCCCGGTACGCCTGGCGGCACCAGGAG CTGTGGCGGGAGCCCACTCCCCCGGTGGCCGCCGGCGGGCACGACATCGGCTCGCACACC CTGCTCGGCACGGCGACCACCGTCGCCGGCAGCGACCTGCGGCTGTGGCACACGGTGCTC AGCGACGCCAGCCGTCCGTACCCGGGCAGCCACACCGTGCAGGGCGCGGAGCTCGTGCCG GCCGCGGTGCTCGCCGCCACCTTCCTCGCCGCCGGGGCCCGCGACGGCGCGCCGGTGGCC CTGCACGACCTGTCGATGCGGGTGCCGCTGGCCACGGCCGGGCGCCAGGAGATCCAGGTG GTGGACGACGGCGGGAAGCTGCTGCTCGCCTCCCGCCCGGCCGGCGCGGACACCGCGCCG TGGCTGACCCACGCCACCGCGCGCGTGACGCCGGACGACGCCGCCCCGTCGGCGCGGGCC GCCGACCCCCGGGGGCCGGGCGGGGTGGTCGCCGACCCGGGGCTCGTCCGGGACCGGCTG GCCCGGGTCGGGGTGCCCGAGACGGGCTTCCCATGGCGGGTCGACCGCCTCACCACCGGC AACGGCGGGCTGCGGGCCCGGGTCCGCTTCCCCGAGCCGGCCGAGGACTGGGCGGCCGTC ATCGACGCGGCGGTGTCCATCGCGCCGGCCGCGTTCCCCGGCGAGCCCCGGCTGCGCCTG GTCGACGCGATCGAGCGGATCTGGACGACCGGCGTGGTGCCGGCCGTCGCCCTCGTCGAC GTCGTCCGGGACGGCGGGCGGGAGGACACCGTCGACGTGACGGTCTCCGGGGCGGACGGC ACTCTCGCCGCCCGGCTCACGGGCCTGCACTACCCGGTGGTCCCCGACGCCCCGGCCGCC GACACCGACGGGCCCGCCCCGACCCGGCAGTCCCTGGCCGACCTCGACCCGGACGAACTG CGCGAGCGCCTGATCGAGGAGGTCCGCGCGGCGGTCGCCGCCGAGATGAAGCTGGCCGCC GAGGCGCTCGACCCGCGCCTGCCGCTGGTGCAGCAGGGCATGGACTCGGTCATGACCGTC ATCGTGCGCCGGCGGCTGGAGAAGACCTACCGCCAGGTGCTGCCCGCCTCGTTGTTCTGG CAGCAGCCCACCGTCACGGCGATCGCGGTGCACCTGGCCGAGCTGATCGCCGCCCCCTCG CCGGACGGGGCGGTCGCCGACGCCGTGGCCCGCTGA
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR001227 Acyl transferase domain (Domain)
IPR009081 Acyl carrier protein-like (Domain)
IPR014030 Beta-ketoacyl synthase, N-terminal (Domain)
IPR014031 Beta-ketoacyl synthase, C-terminal (Domain)
IPR014043 Acyl transferase (Domain)
IPR016035 Acyl transferase/acyl hydrolase/lysophospholipase (Domain)
IPR016036 Malonyl-CoA ACP transacylase, ACP-binding (Domain)
IPR016038 Thiolase-like, subgroup (Domain)
IPR016039 Thiolase-like (Domain)
IPR018201 Beta-ketoacyl synthase, active site (Active_site)
IPR020801 Polyketide synthase, acyl transferase domain (Domain)
IPR020806 Polyketide synthase, phosphopantetheine-binding domain (Domain)
IPR020841 Polyketide synthase, beta-ketoacyl synthase domain (Domain)
|
| SignalP | No significant hit |
| TMHMM | No significant hit |


 Calm_00630
Calm_00630


