Sphingobium sp. SYK-6 (= NBRC 103272)
Lignin is the most abundant aromatic compound in nature. Biodegradation of lignin in lignocellulosic biomass is thought to be initiated by white rot fungi, and the resultant low-molecular-weight products are further degraded and utilized by soil bacteria. Sphingobium sp. SYK-6 is a bacterium isolated from the wastewater of a kraft pulp mill, and is one of the best-characterized
degraders of lignin-derived aromatic compounds, being able to utilize various lignin-derived biaryls, including beta-aryl ether, biphenyl, and diarylpropane, as sole sources of carbon and energy.
Genome analysis of Sphingobium sp. SYK-6 revealed one circular chromosome (4,199,332bp, 65.57% G+C, 3,913 ORFs, 50 tRNAs, 2 rRNA operons) and one circular plasmid pSLGP (148,801bp, 64.40% G+C, 150 ORFs). Genes known to be involved in the degradation of lignin-derived aromatic compounds were located in at least 10 different loci scattered across the chromosome. In addition, many
transporter genes possibly involved in the uptake of a variety of aromatic compounds were predicted. Comparison with Sphingobium japonicum UT26S, whose genome sequence has previously been determined, about 56% of total ORFs in Sphingobium sp. SYK-6 were orthologous to those of Sphingobium japonicum UT26S. Despite the poor conservation of synteny between these two
strains, about 120 ORFs on the plasmid pSLGP were found to be almost identical to those on a chromosomal region of Sphingobium japonicum UT26S, suggesting the plasmid-mediated gene transfer between these sphingomonads.
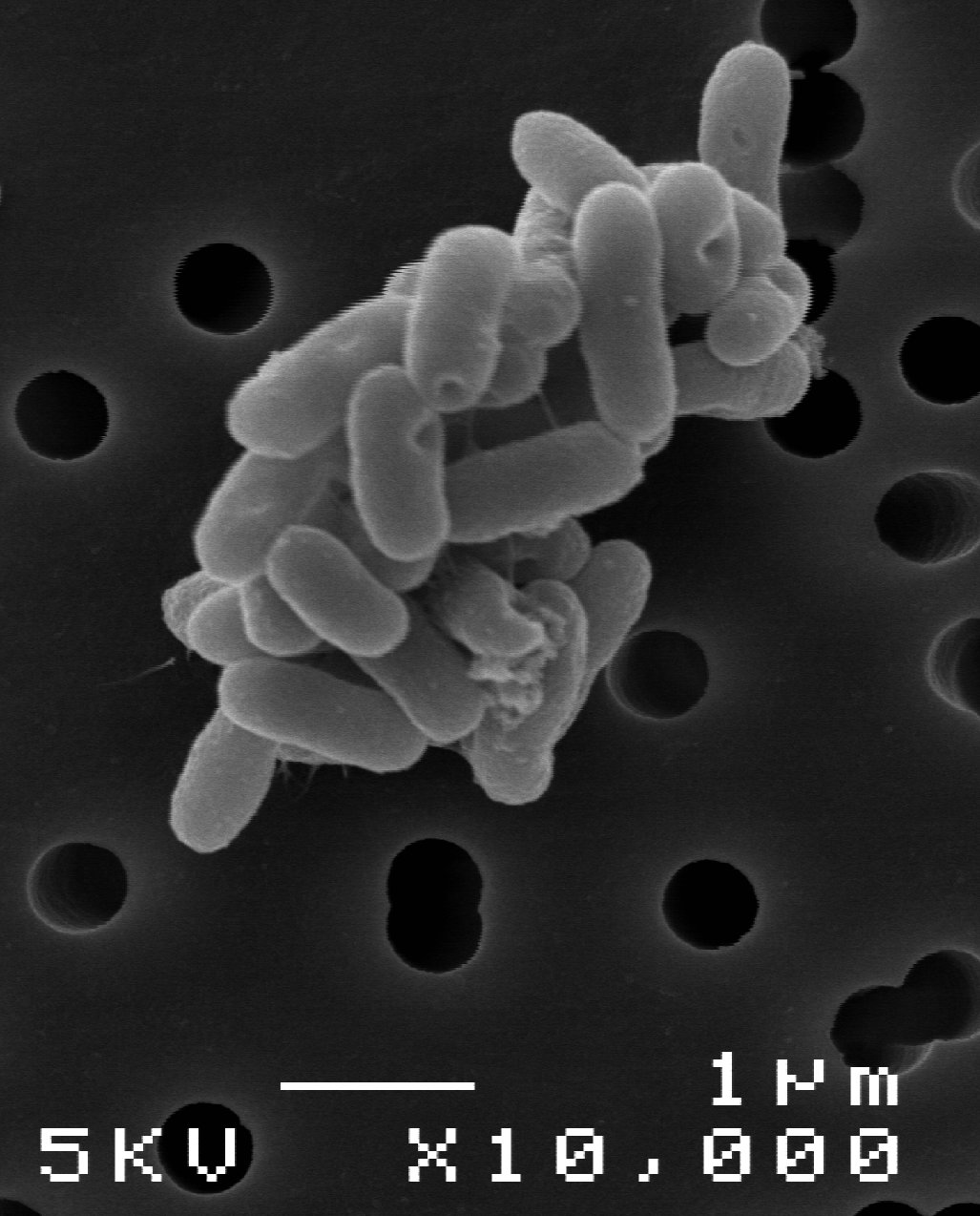
Photo by Nagaoka University of Technology
References:
- [1] Degradation of 3-O-methylgallate in Sphingomonas paucimobilis SYK-6 by pathways involving protocatechuate 4,5-dioxygenase.
- Kasai D.,Masai E.,Katayama Y.,Fukuda M. (2007)
FEMS Microbiol. 274:323-8. [PMID:17645527] - [2] Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds.
- Masai E.,Katayama Y.,Fukuda M.(2007)
Biosci. Biotechnol. Biochem. 71:1-15 [PMID:17213657] - [3] Detection and localization of a new enzyme catalyzing the beta-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6).
- Masai E.,Katayama Y.,Nishikawa S.,Yamasaki M.,Morohoshi N.,Haraguchi T.(1989)
FEBS Lett. 249:348-52. [PMID:2737293]
| Genomic size | 4,348,133 bp |
|---|---|
| The number of ORFs | 4,063 |
| GC content | 65.53% |
| Genome Database | DOGAN |
| NBRC* No. | 103272 |
* The URL of NBRC : http://www.nbrc.nite.go.jp/e/index.html.
- Distribution of Our Microbial Genomic DNA
- At the Biological Resource Center of the National Institute of Technology and Evaluation (NITE Biological Resource Center, an Incorporated Administrative Agency), we have been distributing the microbial genomic DNA.
Contact us
- Biotechnology Evaluation and Development Division, Biological Resource Center, National Institute of Technology and Evaluation
-
Address:2-49-10 Nishihara, Shibuya-ku, Tokyo 1510066, Japan MAP
Contact Form


