Rubra_00400 : CDS information
 Location
Location
| Organism | Streptomyces achromogenes var. rubradiris |
|---|---|
| Strain | NRRL 3061 (=NBRC 14000) |
| Entry name | Rubradirin |
| Contig | AJ871581 |
| Start / Stop / Direction | 77,653 / 81,606 / + [in whole cluster] 77,653 / 81,606 / + [in contig] |
| Location | 77653..81606 [in whole cluster] 77653..81606 [in contig] |
| Type | CDS |
| Length | 3,954 bp (1,317 aa) |
 Annotation
Annotation
| Category | 1.4 Other |
|---|---|
| Product | aminocoumarin--carboxylic acid ligase/L-tyrosine--PCP ligase/peptidyl carrier protein amide synthetase/L-tyrosyl-PCP synthetase/peptidyl carrier protein |
| Product (GenBank) | putative CoA ligase |
| Gene | |
| Gene (GenBank) | rubC1 |
| EC number | |
| Keyword |
|
| Note | |
| Note (GenBank) | |
| Reference |
|
| Related Reference |
|
|
 PKS/NRPS Module
PKS/NRPS Module
| B | 1 | 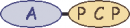 | tyrosine |
|---|
 Sequence
Sequence
>aminocoumarin--carboxylic acid ligase/L-tyrosine--PCP ligase/peptidyl carrier protein [putative CoA ligase] MRGHDHYIRRILGRLDAHPAGVPFVLGNDPFPAERLARAIRTTAGWMRAAGVGPGTSVAV LTSPNTPSTLVYRYAVNLLGATAVHIRGVNAADPQDELGPRVQAEILAGLRPSVLAVDQD NLDRARELRAAAGSAFALAAPGPYGPDVLDMSRPLESPFDDATAERAEIAVVTFTSGSSG RPKGVCWPFDVKDDMASAAAGAQPAVCLITGTLTHSSGFSADDAIIAGGSVVLHHGFDAE AVLRAVERHRVTRLVLASAQVYALTEHHAFDDYDRSSLREVFYTGSPAAPERLAAAAKSL GPVLFQVYGTSETGMISLLTPQDHLDPDLRRTVGRPPANVRITIRDPRDHDRLLPPGVSG EICSSGRWAMSHYWNDPEQTARTVRDGWVRTGDIGRLDESGYLTLEGRLDGVLKGHGVRI HPEAIERVLLEHADVAQAAVFGIEDEDFLALVHAVVTPAPGCTPRPADLGAYVADALGDR HAPVDIEVRSELPLLGSAKPDRNLLREQALGARFWHASLGGVDGDSLLRDRLARPGDDTL GAPALQRRIPDFPSAKNTLLGVWAVLLHLLGAGSPVVFGAGPRPLLVAVAPGEEIGAVLH RVEEAVAASAGHARLGPRGRAELDAAADTLFDTALTFTRVGDPPPGPARPLTAELRVGHD AGDLVLTLVTDPRRIGADCAGRLLEMVLRLLGAFREAPGQPVASLDLLDPATRRQVLQEW NSSRQEQPAATLGRLWQDAADAHAGRPAVEEAGVTTSYAELDRRAGRLAAAIAQAGAAPG RMVALLLPRSLDLITALLATVRTGAAFVPIDPGYPPERITAMLEDCDPVAVCTTASADMP STRPWPRVHLDGPEAGEPAGGFPRVAPDVADPAYVIYTSGTTGRPKGVVVTHAGLANLAA AKREGLGLDSTARVLQFASPSFDAFVAELLGAFTSGATVVVPPQGPLAGEPLTAVLTERR ITHAILPPVALSSMDGAAGALPGLRGLISAGEECPAELAARWSQGRRMVNAYGPTEVTVC ATQSGPLTADGRPPIGRPVAGARAYVLGPGLQPVPPGFRGELYIAGPGVARGYLNQPALT AVRFVADPFGPPGSRMYRTGDIASWRADGNLDFHGRADDQIKLRGFRIEPREVAAVLEEL PAVARAVAGVREDRGGRARLIAWVVPADGEAPTPERLREHAARRLPEHMTPTGYVLVSAL PMTPNGKLDLAALPAVAERDSPPPAAARPALRTPAERALAAIVGELLGATDVGADSDFFA LGGDSMLAISLIRRAREAGLALSPKEIVGNPTIGALAAVATSLPSPPAQTPHVEGRR
>aminocoumarin--carboxylic acid ligase/L-tyrosine--PCP ligase/peptidyl carrier protein [putative CoA ligase] TTGCGGGGGCATGATCACTACATCCGCCGGATCCTCGGCCGGCTCGACGCCCATCCTGCC GGCGTTCCCTTCGTGCTCGGAAACGACCCCTTCCCGGCCGAGCGTCTCGCCCGTGCGATC CGCACGACCGCCGGGTGGATGCGCGCGGCCGGTGTCGGCCCGGGCACCTCGGTCGCCGTG CTGACCAGTCCCAACACACCGTCCACCCTGGTGTACCGGTACGCGGTGAACCTCCTGGGC GCGACGGCGGTCCACATTCGCGGGGTGAACGCCGCGGACCCTCAGGACGAGTTGGGTCCT CGCGTCCAGGCGGAGATTCTCGCCGGGCTGCGGCCGAGCGTGCTGGCGGTGGACCAGGAC AACCTCGACCGTGCGCGCGAGCTGCGTGCCGCGGCCGGATCGGCGTTCGCCCTCGCCGCC CCGGGTCCGTACGGACCGGATGTTCTGGACATGAGCCGCCCCTTGGAGTCGCCGTTCGAC GACGCGACGGCCGAGCGGGCGGAGATCGCCGTGGTGACGTTCACCAGCGGATCGTCGGGA CGGCCCAAGGGCGTCTGCTGGCCGTTCGACGTGAAGGACGACATGGCCTCGGCGGCCGCC GGGGCCCAGCCGGCGGTCTGTCTGATCACCGGGACGCTGACCCACTCCAGCGGTTTCTCG GCCGATGACGCGATCATCGCCGGGGGCAGTGTCGTGCTGCACCACGGGTTCGACGCCGAG GCGGTGCTCCGAGCCGTCGAACGGCACCGCGTCACCCGGCTCGTCCTCGCCTCTGCGCAG GTCTACGCCCTCACCGAGCATCACGCGTTCGACGACTACGACCGCTCCAGCCTGCGCGAG GTGTTCTACACCGGGAGCCCGGCCGCGCCGGAGCGGCTGGCGGCCGCCGCCAAATCGCTC GGCCCGGTGCTGTTCCAGGTCTACGGCACGAGCGAGACCGGCATGATCAGCCTGCTCACC CCACAGGATCATCTGGACCCCGACCTGCGGCGCACGGTCGGCCGGCCCCCGGCGAACGTC CGGATCACCATCCGTGATCCGCGAGACCACGACCGGCTCCTCCCGCCGGGCGTCTCCGGC GAGATCTGCTCGTCCGGGCGCTGGGCGATGAGCCACTACTGGAACGACCCGGAGCAGACC GCGCGGACGGTCCGCGACGGGTGGGTGCGCACCGGCGACATCGGACGGCTCGACGAGTCG GGATACCTGACCCTCGAGGGCCGCCTCGACGGGGTCCTGAAGGGTCACGGTGTCCGCATC CACCCGGAGGCGATCGAGCGCGTGCTCCTCGAACACGCCGACGTGGCCCAGGCCGCGGTG TTCGGGATCGAGGACGAGGATTTCCTGGCGCTGGTCCACGCGGTGGTGACGCCGGCGCCC GGCTGCACGCCGCGGCCGGCCGACCTGGGGGCGTACGTGGCGGACGCGCTCGGTGACCGC CATGCTCCGGTGGACATCGAGGTCAGGTCCGAACTGCCGCTGCTGGGATCGGCGAAGCCG GACCGGAACCTGCTGCGCGAGCAGGCGCTGGGCGCGCGGTTCTGGCACGCGTCGCTAGGG GGGGTCGACGGTGATTCCCTGCTCCGGGACAGGCTCGCCCGGCCGGGCGACGACACGTTG GGGGCACCCGCACTCCAGCGGCGGATCCCGGATTTCCCGTCCGCAAAGAACACGCTGCTC GGAGTCTGGGCGGTCCTGCTGCACCTGCTGGGCGCCGGCTCGCCGGTCGTCTTCGGGGCC GGGCCCCGGCCGCTGCTGGTGGCCGTCGCGCCGGGCGAGGAGATCGGCGCCGTCCTCCAC CGGGTCGAGGAGGCCGTGGCCGCCTCGGCCGGGCACGCCCGGCTGGGACCGCGGGGGCGG GCCGAACTCGACGCCGCGGCAGACACGTTGTTCGACACCGCGCTCACCTTCACCCGTGTC GGGGACCCGCCGCCCGGCCCGGCCCGGCCGCTCACGGCCGAGCTCAGGGTGGGCCACGAC GCGGGCGATCTCGTCCTGACCCTGGTCACCGACCCGCGGCGGATCGGCGCGGACTGCGCC GGACGCCTCCTCGAGATGGTCCTGCGCCTGCTCGGGGCATTTCGCGAGGCGCCCGGCCAG CCGGTCGCCTCGCTGGACCTCCTCGACCCGGCCACCCGCCGCCAGGTGCTGCAGGAATGG AACTCCTCCCGGCAGGAGCAGCCCGCCGCGACCCTGGGCCGGCTGTGGCAGGACGCTGCG GACGCACACGCCGGACGGCCGGCCGTGGAAGAGGCCGGCGTCACCACCTCGTACGCCGAA CTCGACCGCCGTGCCGGCCGGCTCGCGGCGGCCATCGCGCAGGCGGGTGCGGCGCCGGGC CGGATGGTGGCCCTCCTGCTGCCGCGGTCCCTGGACCTGATCACCGCGCTCCTGGCGACC GTCCGGACCGGCGCGGCGTTCGTCCCCATCGACCCGGGATACCCGCCAGAGCGGATCACG GCGATGCTGGAGGACTGCGACCCGGTGGCGGTCTGCACGACGGCGTCGGCCGACATGCCC TCGACCCGTCCCTGGCCGCGCGTCCACCTGGACGGGCCGGAGGCCGGCGAGCCGGCGGGC GGGTTTCCCCGGGTCGCGCCCGACGTGGCCGATCCGGCGTACGTCATCTACACCTCCGGC ACGACCGGCCGGCCGAAGGGCGTCGTGGTCACCCACGCCGGGCTGGCGAACCTGGCCGCG GCCAAACGCGAGGGCCTGGGACTGGACAGCACCGCCCGGGTCCTGCAGTTCGCCTCACCG AGCTTCGACGCGTTCGTCGCCGAGCTGTTGGGCGCGTTCACCTCGGGGGCGACGGTGGTC GTGCCGCCGCAGGGCCCCCTCGCCGGAGAACCGCTGACGGCGGTTCTGACCGAGCGCCGG ATCACCCACGCCATCCTGCCGCCGGTGGCACTGTCCAGCATGGACGGCGCCGCCGGCGCA CTGCCCGGGCTCCGCGGCCTGATCAGCGCGGGCGAGGAGTGCCCGGCCGAGCTGGCCGCG CGCTGGTCGCAGGGCCGCCGGATGGTCAACGCCTACGGGCCCACCGAGGTCACCGTCTGC GCCACTCAGAGCGGGCCGCTCACGGCAGACGGCCGGCCTCCGATCGGACGGCCCGTCGCC GGTGCCCGCGCCTACGTGCTCGGCCCCGGGCTGCAGCCGGTGCCACCCGGATTCCGCGGC GAGCTCTACATCGCGGGCCCCGGCGTCGCCCGGGGTTACCTCAACCAGCCCGCCCTGACA GCCGTACGCTTCGTGGCGGACCCGTTCGGCCCGCCCGGCAGCCGGATGTACCGCACCGGC GACATCGCCTCCTGGCGCGCCGACGGCAACCTGGACTTCCACGGCCGGGCCGACGACCAG ATCAAACTGCGGGGCTTTCGGATCGAGCCGCGCGAGGTGGCGGCGGTGCTGGAGGAACTG CCCGCGGTCGCCCGTGCGGTGGCCGGCGTACGCGAGGACCGTGGCGGCCGGGCCCGGCTG ATCGCCTGGGTCGTGCCCGCCGACGGGGAGGCACCCACCCCCGAGCGGCTCCGGGAGCAC GCGGCACGCCGGCTGCCGGAGCACATGACGCCGACGGGGTACGTCCTCGTCAGTGCCCTG CCCATGACGCCGAACGGCAAGCTGGACCTGGCCGCGCTGCCCGCGGTCGCCGAGCGGGAC AGTCCCCCGCCCGCGGCGGCGCGGCCGGCCCTGCGCACGCCGGCGGAACGCGCGCTGGCC GCCATCGTCGGGGAGCTGCTGGGCGCCACGGACGTCGGCGCCGACAGCGACTTCTTCGCG CTCGGCGGCGACAGCATGCTGGCCATCTCGCTGATCCGGCGCGCCCGCGAAGCGGGTCTG GCCCTCTCCCCCAAGGAGATCGTGGGAAACCCCACGATCGGTGCCCTCGCCGCGGTGGCC ACCTCGCTGCCGTCCCCGCCCGCCCAGACGCCTCACGTGGAAGGCCGCCGATGA
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR000873 AMP-dependent synthetase/ligase (Domain)
IPR009081 Acyl carrier protein-like (Domain)
IPR010071 Amino acid adenylation domain (Domain)
IPR020845 AMP-binding, conserved site (Conserved_site)
|
| SignalP | |
| TMHMM | No significant hit |


 Rubra_00390
Rubra_00390


