Avil_00210 : CDS information
 Location
Location
| Organism | Streptomyces viridochromogenes |
|---|---|
| Strain | Tü57 |
| Entry name | Avilamycin |
| Contig | AF333038 |
| Start / Stop / Direction | 17,621 / 21,502 / + [in whole cluster] 17,621 / 21,502 / + [in contig] |
| Location | 17621..21502 [in whole cluster] 17621..21502 [in contig] |
| Type | CDS |
| Length | 3,882 bp (1,293 aa) |
 Annotation
Annotation
| Category | 1.1 PKS |
|---|---|
| Product | polyketide synthase orsellinic acid synthase |
| Product (GenBank) | polyketide synthase |
| Gene | |
| Gene (GenBank) | aviM |
| EC number | |
| Keyword |
|
| Note | |
| Note (GenBank) |
|
| Reference |
|
| Related Reference |
|
 PKS/NRPS Module
PKS/NRPS Module
| 1 | 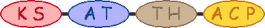 | acetyl-CoA malonyl-CoA |
 Sequence
Sequence
>polyketide synthase [polyketide synthase] MMNTGNDERNMPVISLDRPADSLSPVAVVGIGCRFPGGVNSPGEFWDLLTAGRNTVGEMP PDRWEEYRDFGPRFDAALRTAIRSGSFLDDDIAGFDAEFFGISPREAELMDPQQRLMLEV AWQALEHAGIPPHTLAGTDTGVFAGVCTYDYGAGRLEDLPNIDAWTGIGAAVCAVSNRVS HALDLRGPSLSIDTACSASLVALHTAAQSLRLGECTVALAGGVNLLVSPGQTIALGTAGA LAPDGRSKPFAASAGRYAVAASADGYGRGEGCGVLVIKLLTDAVRDGDRVLAVLRGSAFN QDGRTNGIMAPCGQAQEHVMRRALTAAGVAADTVDFVEAHGTGTRLGDPMEIGAIAAVYG RDRSGQEPCAVGSVKSNIGHLEGAAGVAGVIKAILALDEDRIPASLLDGDPNPEIDWAGL DIRLATRALPWPERPHPRRAAVSGFGYGGTVAHVVLEQAPTAPARPAPEPAGTLFPVSAA SPEALRDRAAALAERVEEGADLASVGHTLAHRQSPLVHRAAVVATGRDELAAGLRALATQ EPAPGLVTGAALPDAGRPVWVFSGHGSQWAGMGRELLEAEPVFAEVIDELEPVFKEEIGF SPRQMLLEGDHTEVDGAQTMIFAMQLGLAALWRSRGVEPAAVIGHSVGEIAAAVTAGALT VTDGARLICRRSLLLREAAGRGAMAMVSLPFDEAAERLAGNDAVVAAIASSTTSTVISGD PGEVEKVVGRWTDEGLVVRRVASDVAFHSPHMDPLLDRLRAAADELSPSAPHTPLYTTAL ADPRATVTADGAYWAANLRNPVRLAAAITAAAEDGHRAFVELSPHPVVAHSIHETLAERG VEDVFVGPTLRRNQPEARTFRAAVGAAHCHGVSVDWSALQPNGNLEVLPPYPWQHRPLWR SIAGARAAERGHDVDSHTLLGTPGGVAGSDLRLWHSTLDDDSRPYPGSHALNGVEIVPAA VLAVTFLAAGAEGEERRALQDMTMTHPVLTAGQRQIQVVREGEVVRLASRTVADAADPNP AWLVHAEARTAAPDLAGLAARSLLDPGEHRLEPADPGLVSRRLAEVGVPSTGFDWSVERL SAGLGVLHAQVLSPDASSWAPLLDAVMSIAPAAFVGLPQLRMVVHVDEITVDGTPPEAAT VEVALDPRVADTVHALVTDGEGRPVASLRGLRYPVVEQPAAPDTDEPGGDADADVVSFAG LSPEELRSRVLDEVREQIAQEMRLAPTALHVRRPLVEQGLDSVMTVVVRRRLEKRLGRDV PANIFWKLPTISDIVDHLTERLTEHPTADGHAS
>polyketide synthase [polyketide synthase] ATGATGAATACCGGCAACGACGAACGGAATATGCCCGTGATATCCCTTGATCGCCCGGCG GACTCCCTCTCTCCGGTGGCCGTGGTCGGGATCGGATGCCGGTTCCCCGGTGGTGTCAAC TCGCCCGGCGAATTCTGGGACCTGTTGACGGCCGGACGCAACACCGTCGGCGAGATGCCC CCCGACCGCTGGGAAGAGTACCGGGACTTCGGTCCGCGGTTCGATGCGGCGCTGCGGACG GCGATTCGCTCCGGCAGTTTCCTGGACGACGACATAGCGGGTTTCGACGCCGAGTTCTTC GGCATCTCGCCGCGCGAGGCCGAGCTGATGGACCCGCAGCAGCGCCTGATGCTGGAGGTG GCCTGGCAGGCACTGGAGCACGCGGGCATCCCGCCGCACACCCTGGCAGGGACCGACACC GGTGTGTTCGCCGGGGTCTGCACCTACGACTATGGCGCCGGACGGCTGGAAGACCTGCCG AACATCGACGCATGGACGGGGATCGGCGCGGCGGTGTGCGCCGTGTCCAACCGCGTCTCT CACGCGCTCGACCTGCGCGGGCCCAGCCTCTCCATCGACACCGCGTGCTCGGCCTCCCTG GTGGCGCTGCACACCGCCGCGCAGAGCCTGCGGCTCGGCGAGTGCACCGTGGCGCTGGCC GGGGGCGTCAACCTGCTGGTGTCACCGGGCCAGACGATCGCCCTGGGCACGGCCGGCGCG CTGGCGCCCGACGGCCGGAGCAAGCCGTTCGCCGCGTCGGCCGGACGCTACGCCGTTGCC GCGTCGGCCGACGGCTACGGCCGCGGCGAGGGCTGCGGCGTGCTGGTCATCAAGCTGCTG ACGGACGCCGTACGCGACGGTGACCGGGTGCTGGCGGTGCTGCGCGGCAGCGCGTTCAAC CAGGACGGGCGCACCAACGGCATCATGGCCCCCTGCGGGCAGGCCCAGGAGCACGTCATG AGGCGCGCCCTGACGGCCGCGGGCGTCGCTGCCGACACCGTTGACTTCGTCGAGGCCCAC GGCACCGGTACCCGCCTCGGCGACCCCATGGAGATCGGCGCGATCGCCGCCGTCTACGGA CGTGACCGCTCCGGTCAGGAGCCGTGCGCGGTCGGGTCGGTGAAGTCCAACATCGGGCAT CTGGAGGGGGCCGCCGGGGTCGCCGGGGTCATCAAGGCGATCCTCGCCCTCGACGAGGAC CGGATCCCGGCCAGCCTGCTGGACGGCGACCCGAACCCCGAGATCGACTGGGCCGGCCTC GACATCCGGCTGGCGACCCGGGCCCTGCCGTGGCCCGAGCGTCCGCACCCCCGCCGGGCG GCTGTGTCCGGTTTCGGCTACGGCGGCACCGTGGCCCATGTGGTCCTCGAACAGGCGCCC ACCGCGCCCGCCCGCCCCGCGCCGGAGCCGGCCGGAACCCTGTTCCCGGTGTCCGCGGCC TCCCCGGAGGCGCTCCGTGACCGCGCGGCCGCGCTCGCCGAGCGGGTCGAGGAGGGCGCT GACCTGGCCTCGGTCGGGCACACCCTGGCCCATCGGCAGTCCCCGCTGGTCCACCGGGCG GCCGTCGTGGCGACCGGCCGGGACGAACTGGCCGCCGGGCTGCGCGCGCTGGCCACTCAG GAACCCGCGCCCGGTCTCGTCACCGGGGCCGCGCTGCCGGATGCCGGCCGTCCCGTCTGG GTGTTCTCGGGTCACGGGTCCCAGTGGGCCGGGATGGGCCGCGAACTGCTGGAAGCCGAG CCGGTCTTCGCCGAGGTGATCGACGAACTGGAGCCGGTGTTCAAGGAGGAGATCGGGTTC TCGCCCCGGCAGATGCTGCTGGAGGGCGACCACACCGAGGTCGACGGCGCCCAGACCATG ATCTTCGCGATGCAGCTCGGGCTCGCCGCGCTGTGGCGGTCACGGGGCGTCGAGCCCGCC GCCGTCATCGGCCACTCGGTCGGTGAGATCGCCGCCGCCGTCACCGCCGGGGCGCTGACC GTGACCGACGGGGCCCGGCTGATCTGCCGTAGGTCGCTGCTGCTGCGCGAGGCCGCGGGG CGTGGCGCGATGGCGATGGTCAGTCTGCCCTTCGACGAGGCCGCCGAGCGGCTCGCGGGC AATGACGCGGTGGTCGCGGCCATCGCGTCCTCGACGACGTCCACCGTGATCTCCGGTGAC CCCGGCGAGGTCGAGAAGGTCGTCGGCCGCTGGACCGACGAGGGGCTGGTCGTACGGCGG GTCGCCTCCGACGTGGCCTTCCACAGCCCGCACATGGACCCCCTGCTCGACCGGCTGCGC GCGGCCGCCGACGAGCTGAGTCCCAGCGCACCGCACACGCCGCTCTACACGACGGCGCTC GCGGACCCGCGGGCCACGGTGACCGCCGACGGCGCGTACTGGGCGGCGAACCTGCGCAAC CCGGTGCGGCTCGCCGCCGCGATCACCGCGGCGGCCGAGGACGGCCACCGGGCGTTCGTC GAGCTGTCCCCGCACCCCGTGGTCGCGCACTCGATCCACGAGACGCTGGCCGAACGCGGC GTGGAGGACGTGTTCGTCGGACCGACGCTACGGCGCAACCAGCCCGAGGCCCGGACCTTC CGCGCCGCCGTGGGCGCCGCCCACTGCCACGGCGTGAGCGTGGACTGGTCGGCGCTCCAG CCGAACGGGAACCTCGAAGTCCTGCCGCCCTACCCCTGGCAGCACCGCCCGCTGTGGCGT TCCATCGCCGGGGCGCGAGCGGCCGAGCGCGGCCACGACGTCGACTCCCACACGCTGCTG GGCACGCCCGGCGGCGTCGCGGGCAGCGACCTGCGGCTGTGGCACAGCACGCTGGACGAC GACAGCCGCCCGTACCCGGGCAGCCACGCCCTCAACGGCGTGGAGATCGTCCCGGCCGCC GTGCTGGCGGTCACGTTCCTGGCGGCCGGCGCCGAGGGCGAAGAGCGCCGCGCCCTCCAG GACATGACGATGACCCACCCGGTGCTGACGGCGGGTCAGCGGCAGATCCAGGTCGTCCGC GAGGGCGAGGTGGTGCGGCTGGCCTCCCGGACGGTCGCGGACGCCGCCGACCCGAACCCC GCCTGGCTCGTCCATGCCGAGGCCCGGACGGCCGCGCCGGACCTCGCCGGTCTGGCGGCG CGGTCGCTGCTGGACCCGGGCGAGCACCGGCTCGAACCGGCCGACCCCGGCCTGGTCTCC CGGCGGCTGGCCGAGGTGGGCGTACCCTCGACCGGTTTCGATTGGAGCGTCGAGCGACTG TCCGCCGGTCTCGGTGTACTGCACGCTCAGGTGCTCTCGCCCGACGCCTCGTCCTGGGCC CCGCTGCTGGACGCCGTGATGTCGATCGCGCCGGCCGCCTTCGTGGGCCTCCCGCAGCTC CGCATGGTCGTGCACGTCGACGAGATCACCGTCGACGGCACGCCACCGGAGGCGGCGACG GTCGAGGTCGCGCTCGATCCCCGCGTCGCCGACACCGTGCACGCCCTGGTCACGGACGGG GAGGGACGCCCGGTGGCGAGCCTGCGCGGCCTGCGCTACCCGGTGGTCGAGCAGCCGGCC GCGCCGGACACCGACGAGCCGGGCGGCGACGCGGACGCGGACGTGGTGTCGTTCGCGGGT CTGTCGCCGGAGGAGCTGCGTTCCCGGGTGCTCGACGAGGTGCGCGAGCAGATCGCGCAG GAGATGCGACTCGCCCCCACGGCCCTGCACGTTCGCCGCCCGCTGGTGGAGCAGGGGCTC GACTCGGTGATGACGGTGGTGGTCCGCCGCCGGCTGGAGAAGCGCCTCGGCCGGGACGTG CCGGCCAACATCTTCTGGAAGCTGCCCACCATCAGCGACATCGTCGATCACCTGACCGAA CGCCTCACGGAACACCCGACGGCGGACGGCCATGCGTCCTGA
 Feature
Feature
| BLASTP Database:UniProtKB:2011_09 |
|
|---|---|
| InterPro Database:interpro:38.0 |
IPR001227 Acyl transferase domain (Domain)
IPR009081 Acyl carrier protein-like (Domain)
IPR014030 Beta-ketoacyl synthase, N-terminal (Domain)
IPR014031 Beta-ketoacyl synthase, C-terminal (Domain)
IPR014043 Acyl transferase (Domain)
IPR016035 Acyl transferase/acyl hydrolase/lysophospholipase (Domain)
IPR016036 Malonyl-CoA ACP transacylase, ACP-binding (Domain)
IPR016038 Thiolase-like, subgroup (Domain)
IPR016039 Thiolase-like (Domain)
IPR018201 Beta-ketoacyl synthase, active site (Active_site)
IPR020801 Polyketide synthase, acyl transferase domain (Domain)
IPR020806 Polyketide synthase, phosphopantetheine-binding domain (Domain)
IPR020841 Polyketide synthase, beta-ketoacyl synthase domain (Domain)
|
| SignalP | |
| TMHMM | No significant hit |


 Avil_00200
Avil_00200


